FIG 9.
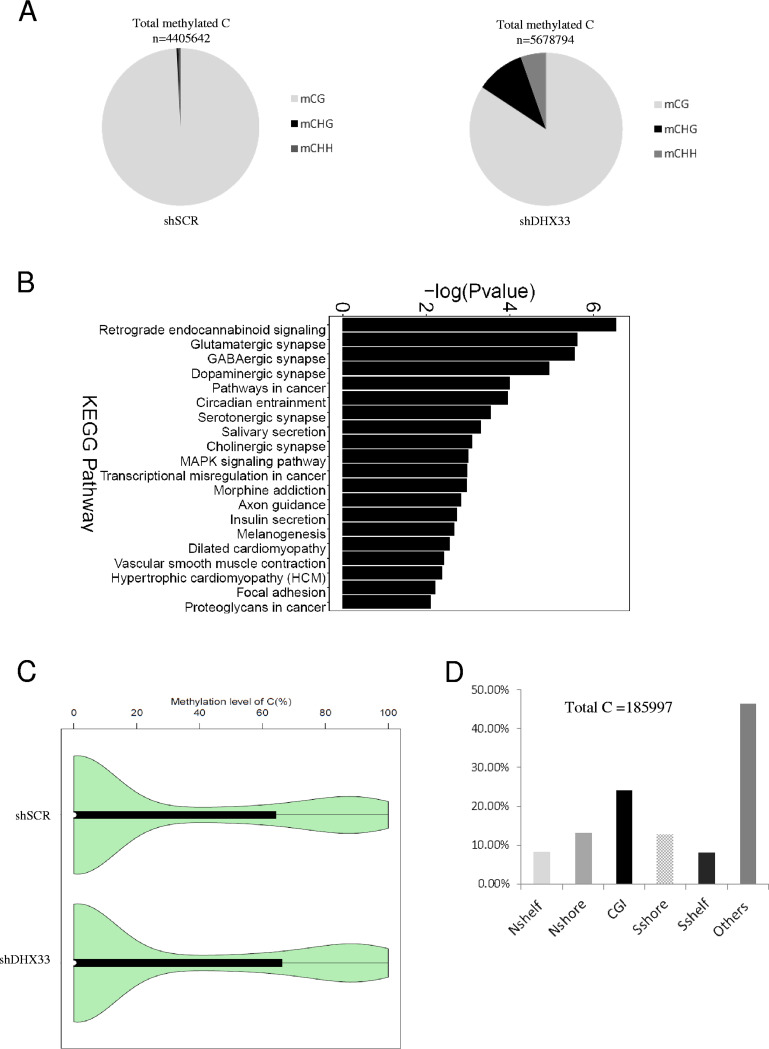
DHX33 deficiency altered epigenetic marks on 5-C at the genomic level. (A) H1299 cells were infected by lentivirus encoding either shSCR or shDHX33. Five days postinfection, cells were subjected to targeted bisulfite sequencing. An SeqCap Epi enrichment system was used to evaluate the epigenetic marks in both shSCR and shDHX33 samples. The composition of methylated C in mCG, mCHG, and mCHH is shown with total methylation sites on the top. (B) KEGG pathway analysis for differentially methylated genes between the control and DHX33 knockdown groups. MAPK, mitogen-activated protein kinase. (C) Violin graph evaluating the methylation levels between the control and the DHX33 knockdown groups. (D) For differentially methylated sites (total, 185,997), results of analyses of their methylation sites are shown in the bar graph. Among them, CGI takes 24.34%.