Figure 1.
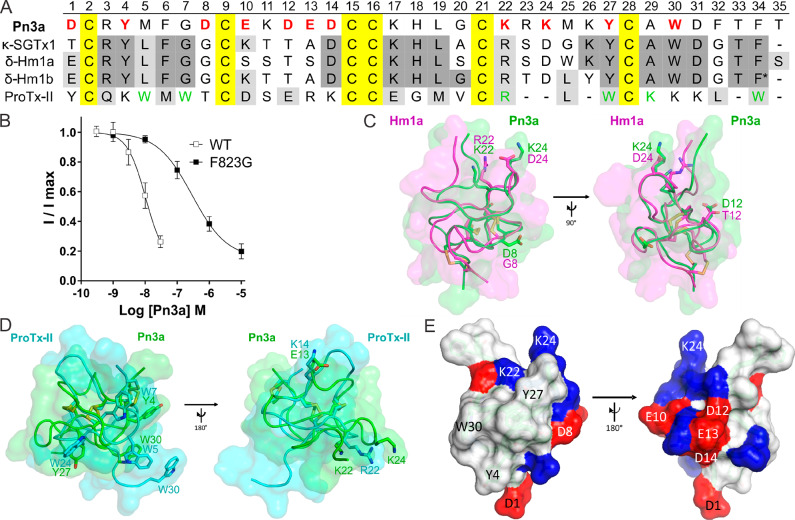
Sequence and structural features of Pn3a (A) Amino acid sequence alignment of Pn3a with closely related venom peptides κ-SGTx1, δ-Hm1a, δ-Hm1b, and ProTx-II. Yellow shading indicates cysteines; dark gray and light gray shading represent identical and similar amino acids, respectively, compared to Pn3a. Red bold letters indicate residues included in SAR study; green letters indicate pharmacophore residues in ProTx-II for NaV1.7 activity.12,30,31 * indicates amidated C-terminus of Hm1b. (B) Pn3a is 28-fold more potent at wild-type mNaV1.7 (IC50 10.4 nM) compared to mNaV1.7[F823G] (IC50 293.4 nM) mutant channels assessed by whole-cell patch–clamp experiments. Data are presented as mean ± SEM, with n = 5–9 cells per data point. (C) Comparison of NMR structures of Pn3a (Protein Databank (PDB) 5T4R; green) with Hm1a (PDB 2N6O; pink) superimposed over the disulfide bonds (yellow), generated with PyMol. Residues of interest are labeled and shown in stick representation. (D) Comparison of NMR structures of Pn3a (PDB 5T4R; green) with ProTx-II (PDB 2N9T; cyan) superimposed over the disulfide bonds (yellow). Overlapping residues with similar chemical properties are labeled and shown as sticks. (E) Surface structure of Pn3a with acidic and basic residues highlighted in red and blue, respectively. The hydrophobic patch (including W30, Y4, and Y27) is surrounded by a charged ring (including D1, D8, K22, and K24; left), while the reverse side is hydrophilic with mainly charged side chains (right). Only residues included in this SAR study are labeled.