Fig. 1.
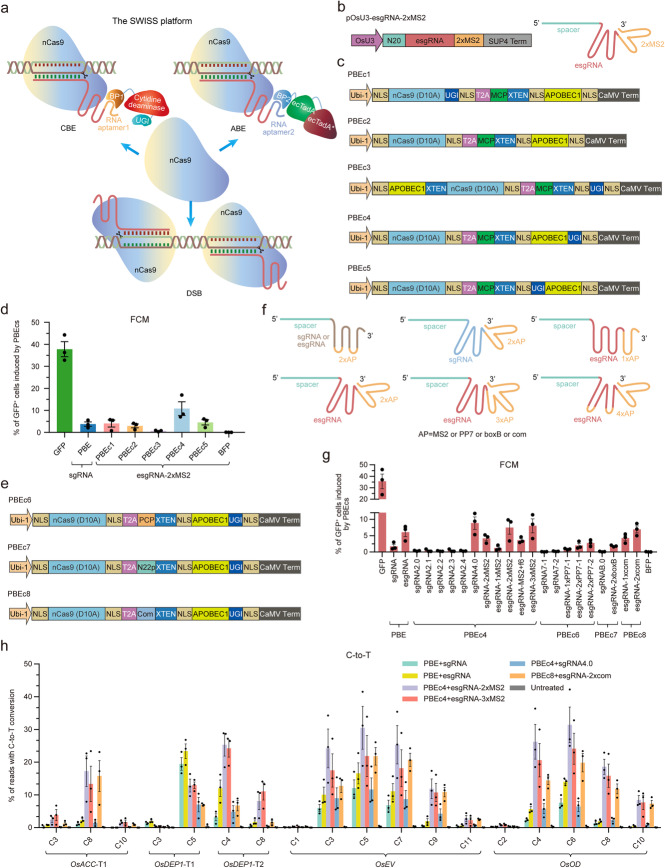
Multiple RNA scaffolds and binding protein orthologs mediate efficient C-to-T conversion. a The CRISPR scaffold RNA-programmed multiplex genome editing system based on nCas9 nuclease. Abbreviation: BP, binding protein. b Architecture of the pOsU3-esgRNA-2×MS2 construct with two MS2 hairpins at the 3′-end of the esgRNA. Abbreviation: SUP4 Term, transcription terminator for the S. cerevisiae SUP4 tRNA gene. c Architectures of PBEc1-c5. Abbreviations: XTEN, 16-aa linker; NLS, nuclear localization signal; CaMV, cauliflower mosaic virus; Term, terminator. d Comparison of C-to-T conversion using a BFP-to-GFP reporter system by PBE and the five PBEcs in rice protoplasts (n = 3). Values and error bars indicate means ± s.e.m. of three independent experiments. e Architectures of PBEc6-c8. Abbreviations: XTEN, a 16-aa linker; NLS, nuclear localization signal; CaMV, cauliflower mosaic virus; Term, terminator. f Schematic of the scRNAs with MS2, PP7, boxB, or com RNA hairpins in the tetraloop and stem loop2 or the 3′-end of the sgRNA and esgRNA. g Comparison of C-to-T conversion using the BFP-to-GFP reporter system induced by various scRNAs and their cognate PBEcs in rice protoplasts (n = 3). The f6 aptamer hairpin binds MCP specifically. Two PP7 hairpin variants were adopted. Values and error bars indicate means ± s.e.m. of three independent experiments. h Comparison of C-to-T editing frequencies of rice endogenous genes induced by four scRNAs and their cognate PBEcs (n = 3). An untreated protoplast sample served as control. Values and error bars indicate means ± s.e.m. of three independent experiments.