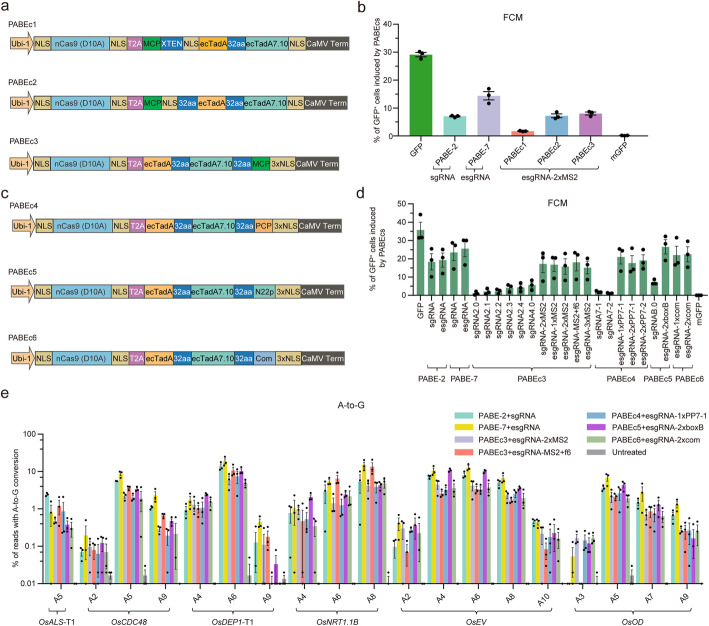
Fig. 2.
Optimization of plant adenine base editor constructs using multiple RNA scaffolds and binding protein orthologs. a Architectures of PABEc1-c3. Abbreviations: ecTadA7.10, evolved Escherichia coli TadA; aa, amino acid; XTEN, a 16 aa linker; NLS, nuclear localization signal; CaMV, cauliflower mosaic virus; Term, terminator. b Comparison of A-to-G conversion using the mGFP-to-GFP reporter system induced by PABE and the three PABEcs in rice protoplasts (n = 3). Values and error bars indicate means ± s.e.m. of three independent experiments. c Architectures of PABEc4-c6. Abbreviations: ecTadA7.10, evolved Escherichia coli TadA; aa, amino acid; XTEN, a 16 aa linker; NLS, nuclear localization signal; CaMV, cauliflower mosaic virus; Term, terminator. d Comparison of A-to-G conversion using the mGFP-to-GFP reporter system induced by various scRNAs and their cognate PABEcs in rice protoplasts (n = 3). Values and error bars indicate means ± s.e.m. of three independent experiments. e Comparison of the A-to-G editing frequencies of endogenous rice genes induced by five scRNAs and their cognate PABEcs (n = 3). An untreated protoplast sample served as control. Values and error bars indicate means ± s.e.m. of three independent experiments