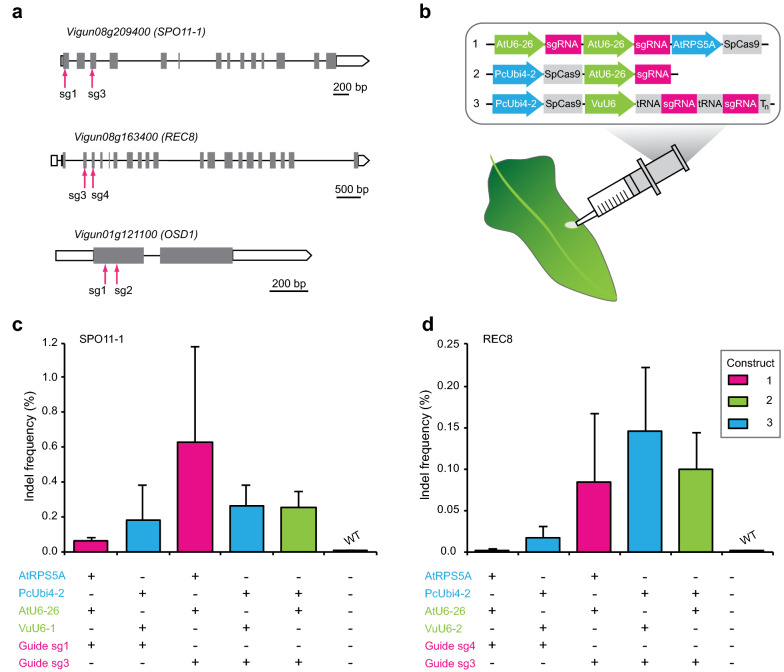
Fig. 3.
Analysis of genome-editing in cowpea leaves infiltrated with CRISPR/Cas9 constructs targeting SPO11-1, REC8 and OSD1.a Cartoons representing structures of SPO11-1, REC8 and OSD1 genes in a 5′-3′ orientation. The CRISPR/Cas9 target sites within each gene are indicated in magenta, exons are indicated by shaded rectangles, introns by intervening lines and untranslated sequences by unshaded rectangles. b Cartoon showing the three types of editing constructs (1–3) tested in the transient assay which differ in promoters driving Cas9 and sgRNA expression (AtRPS5A vs PcUbi4-2 and AtU6-26 vs VuU6, respectively) and the number of guides targeting different genes (two or one). Construct 3 contains two sgRNAs that are expressed as a single polycistronic transcript, driven by VuU6 promoter. T(7) is a 7nt terminator. c Mutagenic efficiency of guides targeting SPO11-1 measured by indel frequency. Each bar represents the % mean value of at least seven independent transient assays ± SE. Non-significant differences from mean values between transiently transformed samples were found within the 95% confidence interval. Information below the X-axis indicates the components of the construct tested in the transient assay. The presence (+) or absence (−) of color-coded construct components match those in the diagram in b. d Mutagenic efficiency of guides targeting REC8 measured by indel frequency. Guides targeting OSD1 did not induce mutations. For further details refer to Additional file 6: Table S9