Fig. 3. Comparison of WT SpCas9 and SpRY nuclease and base editor activities across NNN PAM sites in human cells.
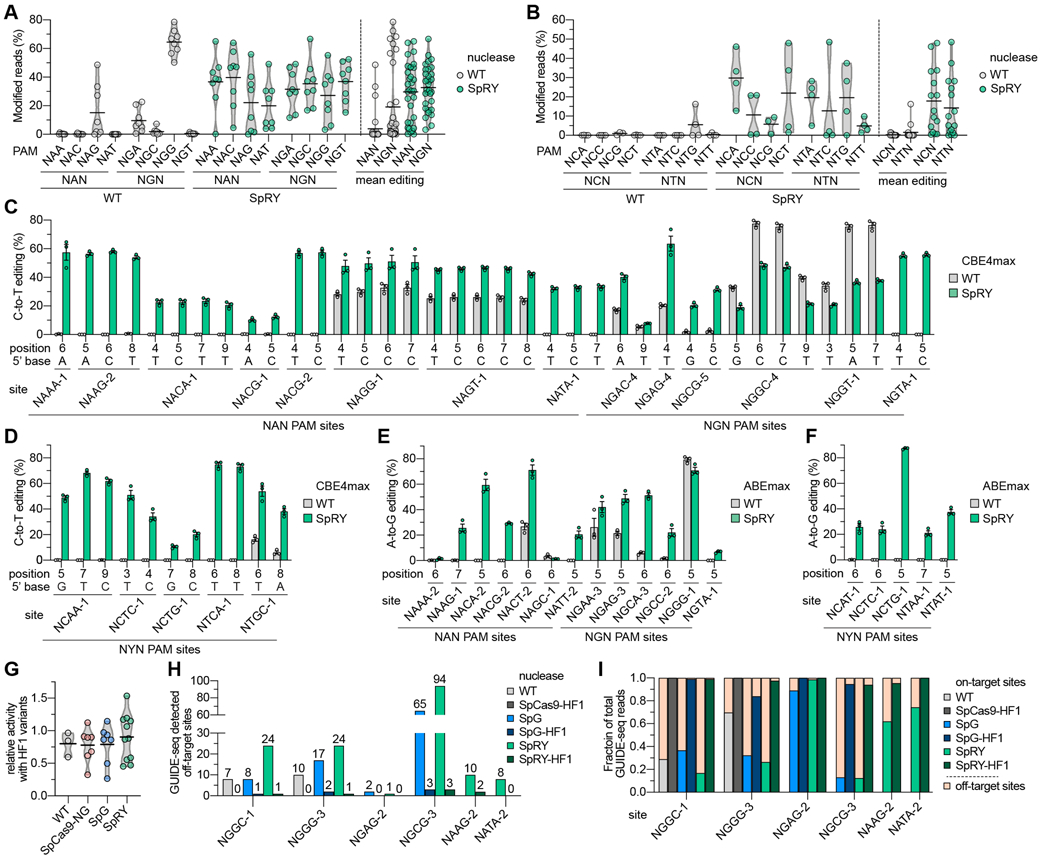
. (A, B) Mean nuclease activity plots for WT SpCas9 and SpRY on 64 sites with NRN PAMs (panel A) and 31 sites with NYN PAMs (panel B) in human cells. The black line represents the mean of 8 or 3-4 sites (panels a and b, respectively) for each PAM of the indicated class (see also Figs. S9A-C), and the grey outline is a violin plot. (C, D) C-to-T base editing of endogenous sites in human cells bearing NRN and NYN PAMs (panels C and D, respectively) with WT SpCas9 and SpRY-CBE4max constructs. Editing of cytosines in the edit window (positions 3 through 9) assessed by targeted sequencing; the five NYN PAM target sites were selected from high-activity sites in panel B; mean, s.e.m., and individual data points shown for n = 3. (E, F) A-to-G base editing of endogenous sites in human cells bearing NRN and NYN PAMs (panels E and F, respectively) with WT SpCas9 and SpRY-ABEmax constructs. Editing of adenines in the edit window assessed by targeted sequencing; the five NYN PAM target sites were selected from high-activity sites in panel B; mean, s.e.m. and individual data points shown for n = 3. For base editing data in panels C-F, see also Table S5. (G) Relative nuclease activity plots for SpCas9-HF1, SpCas9-NG-HF1, SpG-HF1, and SpRY-HF1 compared to their parental variants across 3-10 sites endogenous sites in HEK 293T cells. Mean modification from sites in Fig. S11A shown as dots, with black line representing the mean of those sites, and the grey outline is a violin plot. The HF1 variants additionally encode N497A, R661A, Q695A, and Q926A substitutions(21). (H) Histogram of the number of GUIDE-seq detected off-target sites for SpCas9 variants across sites with NGG, NGN, and NAN PAMs (see Figs. S12A-C, respectively). (I) Fraction of GUIDE-seq reads attributed to the on- and off-target sites for WT SpCas9, SpG, SpRY, and their respective HF1 variants across 2-6 targets (see also Figs. S12A-C and Table S6).
