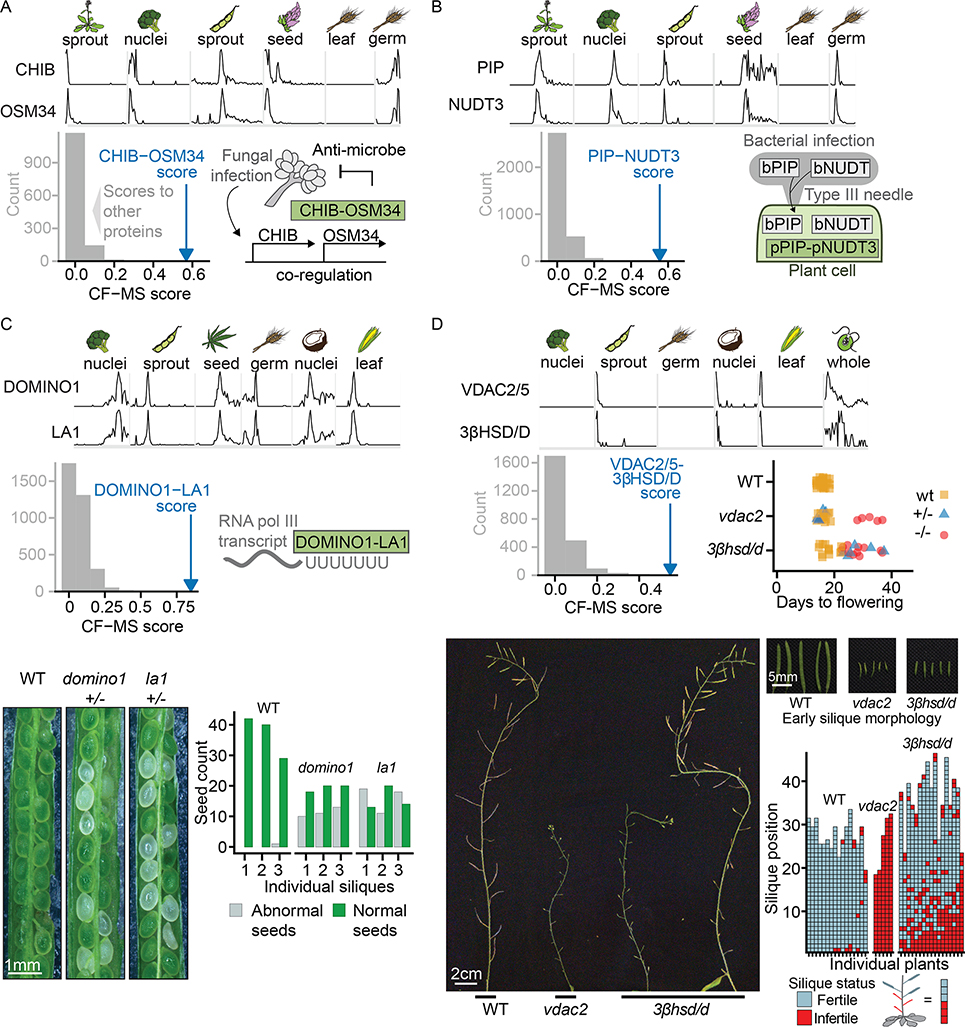
Figure 7. Connecting Plant Genes to Phenotypes via Their Interactions.
The top section of each lettered panel shows sparklines with sample species and tissues indicated above. Bottom left panel of each lettered panel shows the complex interaction score (CF-MS) between subunits (blue arrow) is far greater than the interaction of either subunit with any other observed protein (gray bars representing binned scores).
(A) OSM34 and CHIB form a complex, consistent with co-expression evidence in response to fungal infection (diagram bottom right).
(B) PIP and NUDT3 form a complex in plants. Bacterial members of the PIP and NUDT families are injected into plant cells by a Type III secretion system.
(C) DOMINO1 and LA1 form a plant-specific ribosomal RNA-binding complex and heterozygotes of each have a similar Arabidopsis T-DNA insertion mutant phenotype of abnormal white seeds containing arrested embryos. Bottom left, representative portions of siliques from genotypes as labeled. Lower right, quantification of visually abnormal seeds in three siliques of each genotype. Ratio of normal to abnormal seeds reflects variable penetrance of the mutant phenotype and presence of homozygous and heterozygous embryos in each silique.
(D) Arabidopsis plants homozygous for VDAC2/5 or 3βHSD/D T-DNA insertion mutants show delayed flowering and reduced number of fertile siliques compared to wild type plants of the same stage. Lower panels illustrate fertility defects with main inflorescences at end of flowering. While vdac2 homozygotes produce almost no seeds, 3βhsd/d mutants show a range of fertility levels, ranging from plants with almost no seed-containing siliques to plants in which only early siliques show fertility defects. An enlarged view of wild type and early infertile siliques from plants of the genotypes is shown.