FIGURE 6.
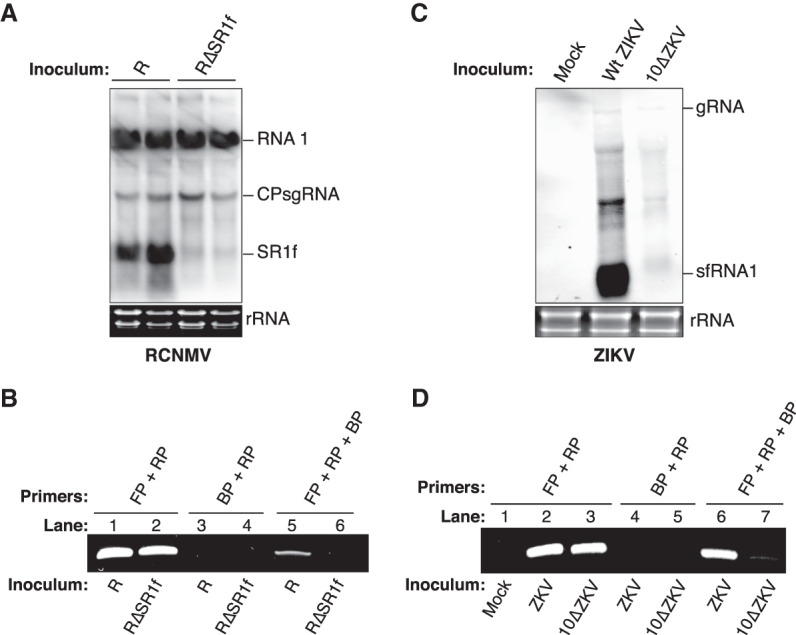
Detection of sgRNAs in virus-infected plants or HeLa cells. (A) Northern blot hybridization of total RNA from N. benthamiana leaves 14 d after inoculation with wild-type (R) or mutant (RΔSR1f) RCNMV. Stained gel shows ribosomal RNA as loading control for each lane. Duplicate samples are shown for each treatment. (B) Detection of SR1f in total RNA from plants infected with indicated wild-type or mutant RCNMV by DeSCo-PCR. Primer combinations used to generate the PCR products are shown above each lane of the gel. (C) Northern blot hybridization of total RNA from HeLa cells 48 h after inoculation with wild-type (Wt) or mutant 10ΔZIKV. Stained gel shows ribosomal RNA as loading control for each lane. (D) Detection of sfRNA1 in total RNA from cells infected with indicated wild-type or mutant ZIKV by DeSCo-PCR. Primer combinations used to generate the PCR products are shown above each lane of the gel. (FP) forward primer, (RP) reverse primer, (BP) blocking primer.
