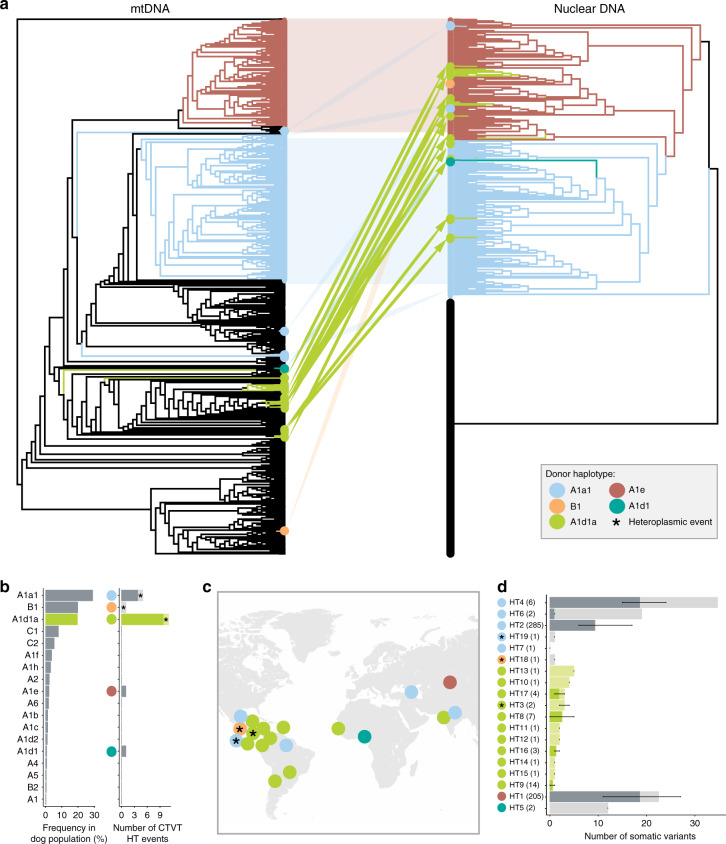
Fig. 1. Recurrent and recent horizontal transfer of canine mitochondrial haplotype A1d1a.
a MtDNA (left) and nuclear DNA (right) maximum likelihood phylogenetic trees for 539 CTVT tumours (coloured) and 494 dogs (black). Correspondence between equivalent CTVTs on mtDNA and nuclear trees is indicated and coloured by mtDNA donor haplotype. Trees are presented as cladograms without informative branch lengths. High resolution trees are presented in Supplementary Figs. 1 and 2. b Frequencies of 18 canine mtDNA haplotypes within a representative global CTVT host dog population (n = 495) (left), and the number of CTVT horizontal transfer (HT) events involving each haplotype in a population of 539 CTVTs (right). Donor haplotype (coloured dots) and heteroplasmic horizontal transfer events (asterisks (*) and lighter shading) are shown. Heteroplasmic tumours carry both the parental and introduced haplotype. The A1d1a heteroplasmic horizontal transfer event involves mtDNA recombination. Bars representing A1d1a are highlighted in green. c Inferred geographical locations of 19 CTVT mtDNA horizontal transfer events. Each horizontal transfer is represented by a dot coloured by donor haplotype. If all CTVTs arising from a horizontal transfer were sampled at the same location, then this was inferred as the location of the horizontal transfer. If CTVTs derived from the horizontal transfer were found in several locations, then the likely site of the horizontal transfer was inferred based on phylogenetic information12,13. Heteroplasmic horizontal transfer events are indicated with an asterisk (*). d Number of somatic mtDNA mutations acquired since each horizontal transfer (HT) event. Number of CTVTs (n = 539) belonging to each HT event is indicated. Bars are split into two categories: darker colour shades represent confident somatic mutations that are polymorphic within each HT group, with error bars representing the mutation range; lighter colour shades represent variants that are fixed within each HT group, whose somatic or germline status cannot be determined (see Methods). Bars representing A1d1a HTs are highlighted in green. Donor haplotype (coloured dots) and heteroplasmy (asterisk, *) are shown.