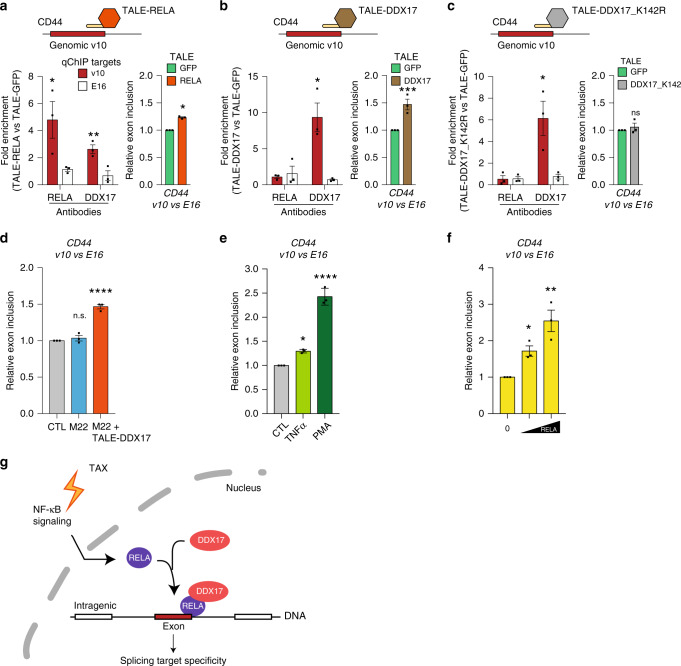
Fig. 6. Chromatin relationship between RELA and DDX17.
a–c Chromatin and splicing regulation upon TALE-mediated tethering of RELA and DDX17. The TALE domain was designed to bind the v10 exon of CD44 and fused to either GFP (a–c), RELA (a), DDX17, (b), or a helicase-deficient mutant DDX17_K142R (c). The effect of TALEs on RELA and DDX17 chromatin enrichment (left panels) and on the relative v10 exon inclusion (right panels) was monitored in HEK cells by qChIP and qRT-PCR, respectively. Results were normalized to measures obtained in TALE-GFP assays. (d) Relative exon inclusion rate of exon v10 of CD44 in HEK cells expressing or not the Tax mutant M22 and the TALE-DDX17 construct. e Relative exon inclusion rate of exon v10 of CD44 in HEK cells exposed to TNFα and PMA. f Relative exon inclusion rate of exon v10 of CD44 in HEK cells transiently transfected with increasing amounts of RELA expression vector (200 and 500 ng). g Model of NF-κB-dependent regulation of alternative splicing. Upon NF-κB activation, DNA-bound RELA proteins act as chromatin anchors for DDX17, which then provides splicing target specificity due to its RNA helicase activity. Data are presented as the mean ± SEM values from biological replicates. Each black square represents a biological replicate. Statistical significance was determined with two-tailed unpaired t-test (qChIP in a–c) and one-way ANOVA followed by Fisher’s LSD test (relative exon inclusion (REI) in a–f) (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). In a–c, exact p-values for TALE-RELA vs TALE-GFP on V10: 0.049 (qChiP RELA), 0.0079 (qChIP DDX17), 0.0276 (REI). Exact p-values for TALE-DDX17 vs TALE-GFP on V10: 0.0139 (qChIP DDX17), 0.0276 (REI). Exact p-values for TALE-DDX17_K142R vs TALE-GFP on V10: 0.0312 (qChIP DDX17). In d–f, exact p-values are <0.0001 (M22 + TALE-DDX17 (d)), 0.0125 (TNFa (e)), <0.0001 (PMA (e)), 0.0369 (0.2 µg (f)), 0.0012 (0.5 µg (f)). Source data are provided as a Source Data file.