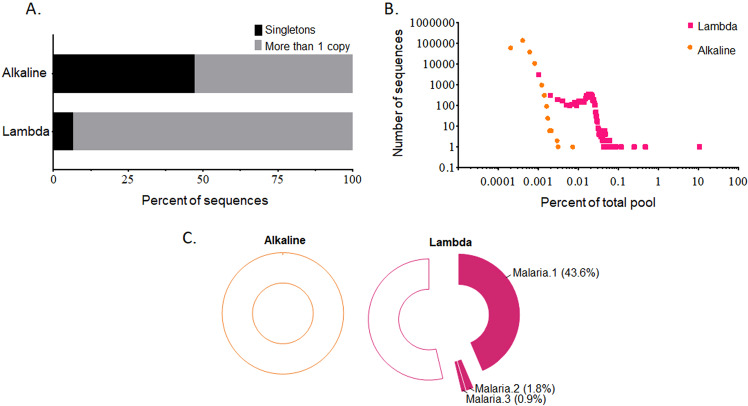
Figure 2.
High-throughput sequencing and bioinformatics analysis of the SELEX enrichment. (A) Based on the percent of individual copies of unique sequences (singletons), sequencing results reveal a reduced enrichment using the alkaline-based SELEX compared with a parallel experiment performed using lambda exonuclease as the ssDNA recovery method. (B) The plot of number of different sequences as a function of enrichment. (C) Representative doughnut graphs of primary sequence alignments. The top 500 unique sequences shows no aptamer clustering from the alkaline SELEX whereas the lambda SELEX reveals a prominent aptamer cluster represented by Malaria.1. Two other clusters, represented by Malaria.2 and Malaria.3 were also identified. Open regions represent sequences that could not be clustered.