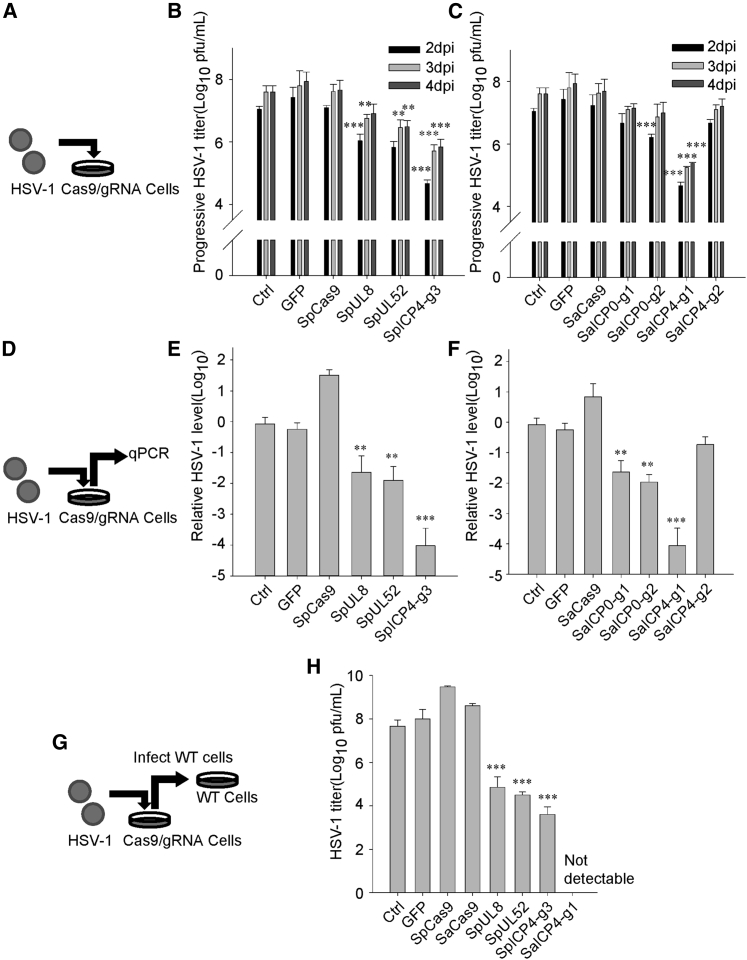
Figure 3.
Expression of SpCas9/gRNA or SaCas9/gRNA Inhibits HSV-1 Replication
(A) Schedule of the experimental process in (B) and (C). HSV-1 was used to infect Cas9 stably expressing Vero cells. (B) Control and SpCas9/gRNA-expressing Vero cells were infected with gradient-diluted HSV-1. Infection progress on each day after infection was determined by the Karber method. (C) Control and SaCas9/gRNA-expressing Vero cells were infected with gradient-diluted HSV-1. Infection progress on each day after infection was determined by the Karber method. (D) Schedule of the experimental process in (E) and (F). HSV-1 (MOI of 0.1) was used to infect Cas9 stably expressing Vero cells. Culture medium containing HSV-1 was collected 3 days post-infection (dpi) and then qPCR was performed to measure HSV-1 genomes. (E) qPCR analysis of HSV-1 genomes in the culture medium of control and SpCas9-expressing Vero cells infected with HSV-1 (MOI of 0.1) at 72 h. (F) qPCR analysis of relative HSV-1 genome copy in the culture medium of control and SaCas9-expressing Vero cells infected with HSV-1 (MOI of 0.1) at 72 h. Data were collected from three independent experiments. Error bars represent standard deviation (SD). Significance was determined by a one-way ANOVA. ∗∗p < 0.01. (G) Schedule of the experimental process in (H). HSV-1 (MOI of 0.1) was used to infect Cas9 stably expressing Vero cells. Culture medium containing HSV-1 was collected at 3 dpi. Viral titer in the medium was measured by the standard Karber method. (H) Titer of HSV-1 in the culture medium of control, SpCas9/gRNA-, or SaCas9/gRNA-expressing Vero cells infected with HSV-1 (MOI of 0.1) at 72 h. Titer was determined by infecting WT Vero cells and calculated by the Karber method on the fifth day after infection. Data were collected from three independent experiments. Error bars represent SD. Significance was determined by a one-way ANOVA. ∗∗p < 0.01, ∗∗∗p < 0.001.