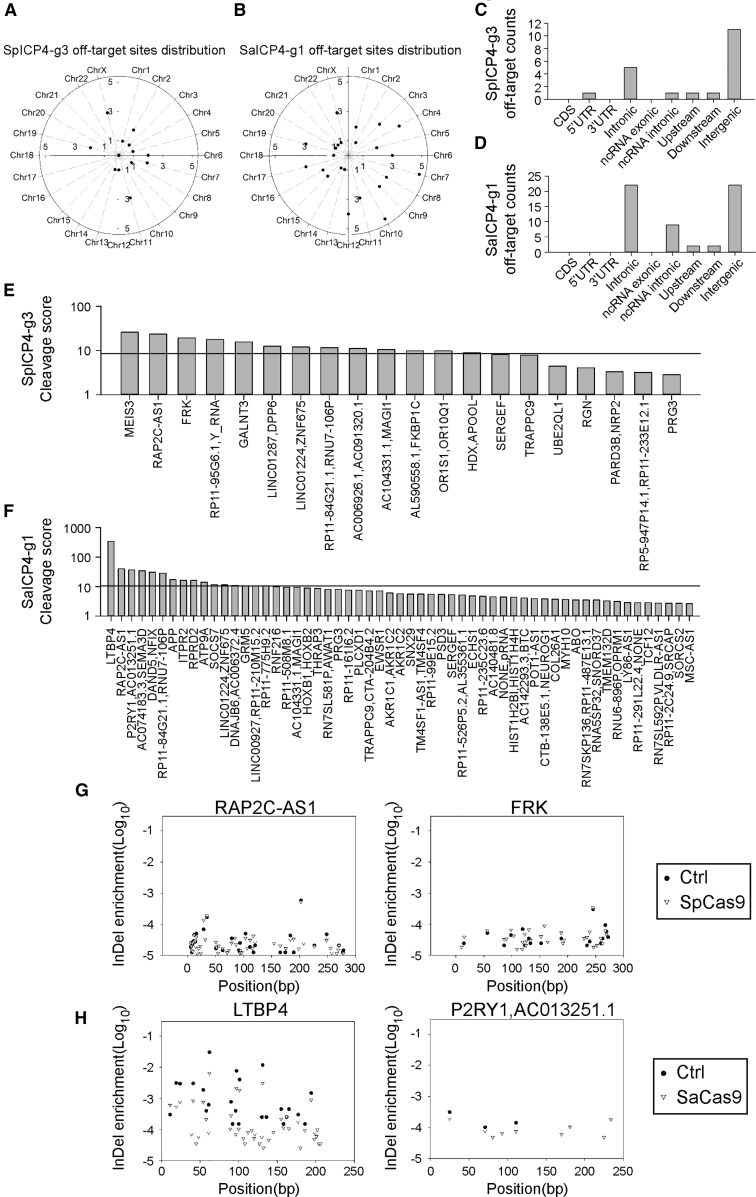
Figure 5.
Digenome Sequencing-Based Off-target Analysis of ICP4-Targeted SpCas9 and SaCas9 gRNAs
(A) Distribution of off-target sites of SpICP4-g3 discovered by digenome sequencing in the whole genome. (B) Distribution of off-target sites of SaICP4-g3 discovered by digenome sequencing in the whole genome. (C) Distribution of off-target sites of SpICP4-g3 discovered by digenome sequencing in different genomic elements. (D) Distribution of off-target sites of SaICP4-g1 discovered by digenome sequencing in different genomic elements. (E) Off-target scores of SpICP4-g3 in each putative off-target site. Black line indicates average score of the control sample. (F) Off-target scores of the SaICP4-g1 gRNA in each putative off-target site. Black line indicates average score of the control sample. (G) Example of deep sequencing confirmation of SpICP4-g3 off-target sites in HEK293T cells. Frequency of indels at a certain position was calculated as the “indel enrichment” score. Scores of control and sample were plotted to identify Cas9 cleavage. (H) Example of deep sequencing confirmation of SaICP4-g1 off-target sites in HEK293T cells. Frequency of indels in a certain position was calculated as the indel enrichment score. Scores of control and sample were plotted to identify Cas9 cleavage.