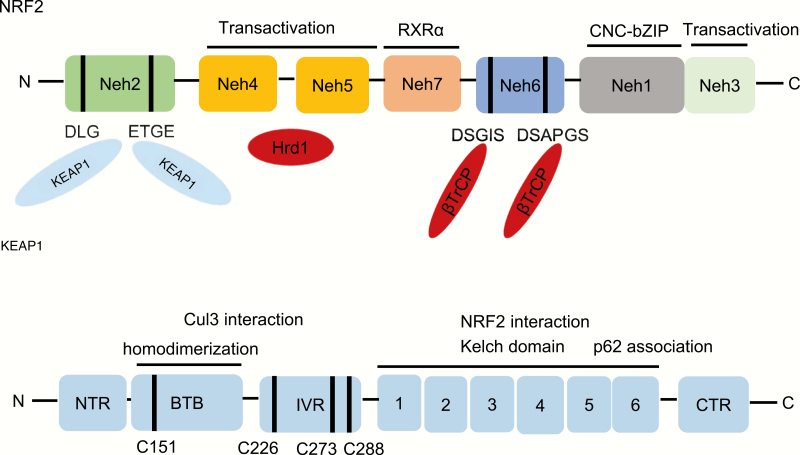
Figure 1.
The architecture of NRF2 and its negative regulator KEAP1. NRF2 contains seven conserved NRF2-ECH homology (Neh) domains, Neh1–Neh7. Neh1 has a bZip motif, where the basic region is responsible for DNA binding and the Zip dimerizes with small MAF proteins. Neh2 contains ETGE and DLG motifs, which are required for binding to KEAP1 and subsequent KEAP1-dependent polyubiquitination and degradation of NRF2. Neh3-5 are transactivation domains and Neh4 and Neh5 domains also interact with Hrd1. Neh6 contains DSGIS and DSAPGS motifs responsible for the β-TrCP-mediated ubiquitination and degradation. The Neh7 domain interacts with RXRα, which inhibits NRF2 activity. KEAP1 contains five domains, an amino terminal region (NTR), a broad complex, tramtrack, bric-a-brac (BTB) domain, an intervening region (IVR), six Kelch motifs and the C-terminal region (CTR). The Kelch motifs and the CTR mediate interactions with NRF2 and p62. The BTB domain mediates homodimerization and contributes to its interaction of the IVR with Cul3. Several functionally important cysteine residues (C151, C226, C273 and C278) sense ROS and electrophiles and are needed in their reduced state for KEAP1 function.