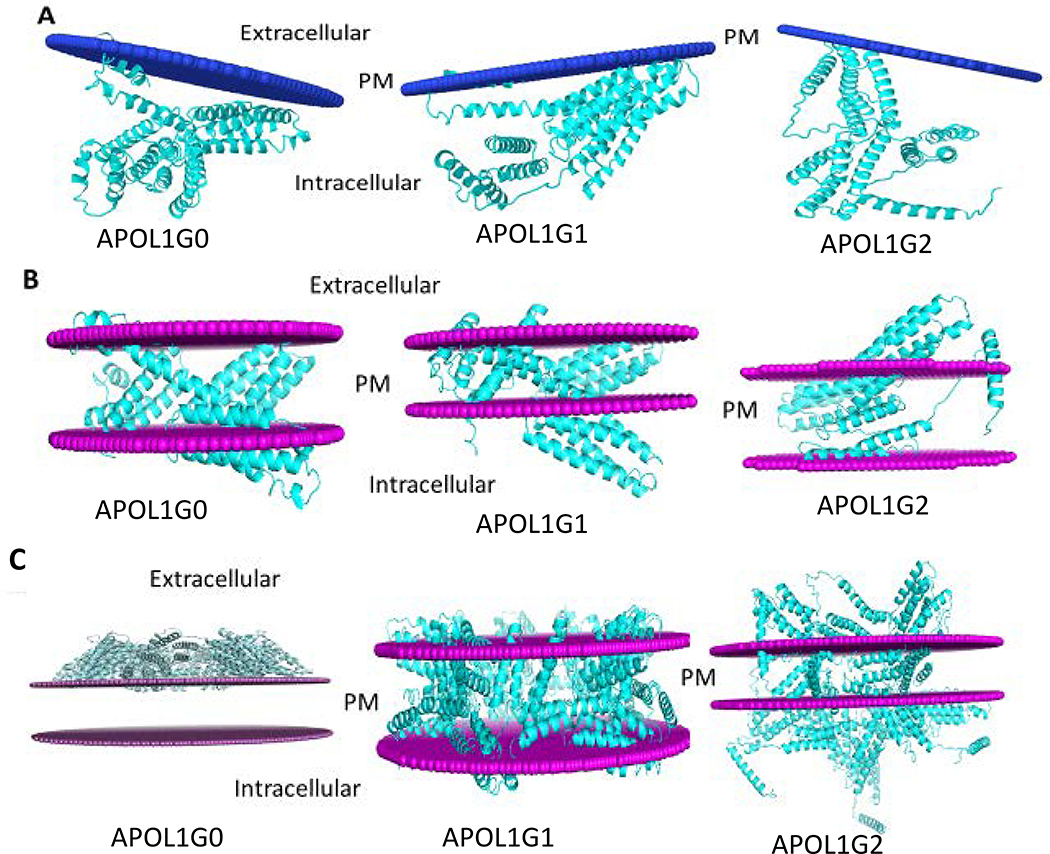
Figure 9. The membrane interaction models for APOL1G0, APOL1G1, and APOL1G2.
A. Peripheral protein model. In this model only membrane interacting helices are attached to the membrane. In APOL1G0, the membrane interacting residues are Ser10, Cys13, Trp15-Ser17 and Leu19-Val23. In APOL1G1, the membrane interacting residues are Glu2, Ala5-Leu6, Val9-Ser10, Leu12-Leu21, Val79, Thr81, Leu86, Trp129 and Gln134. In APOL1G2, the membrane interacting residues are Leu12, Trp15-Ala18 and Trp234.
B. Intgral membrane protein model. In this model APOL1 and its variants have some transmembrane regions. APOL1G0 has 5 transmembrane segments and the hydrophobic thickness is 25.2 Å, APOL1G1 has 7 transmembrane segments and the hydrophobic thickness is 19.8 Å, APOL1G2 has 2 transmembrane segments and the hydrophobic thickness is 33.0 Å.
C. Multimer model of APOL1 variants APOL1G1 and APOL1G2. In this model, APOL1 variants APOL1G1 and APOL1G2 can form multimer in the membrane, however, APOL1G0 multimer is not energetically favorable.