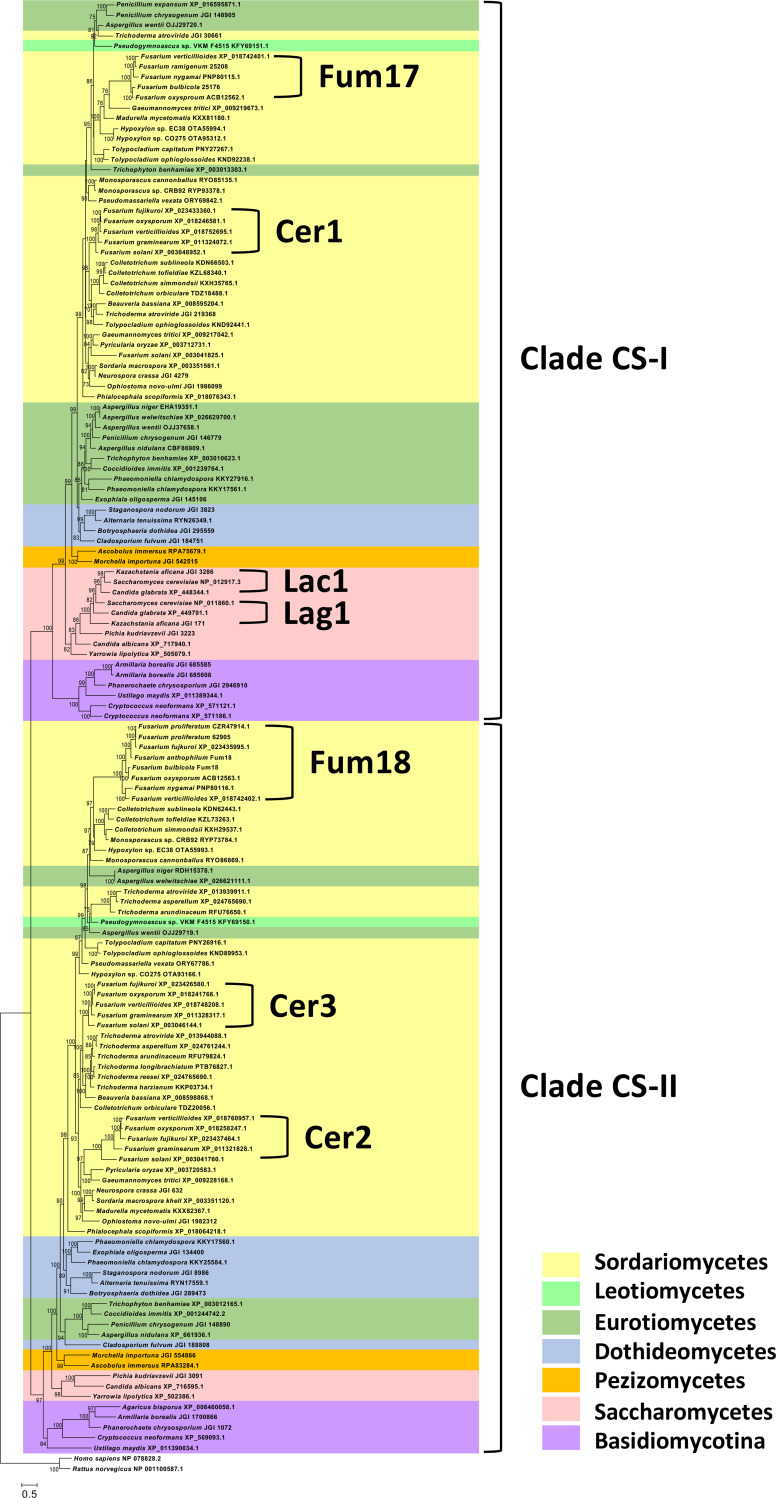
FIG 4.
Phylogenetic tree showing two major clades (CS-I and CS-II) of fungal ceramide synthases and the positions of Fum17 and Fum18 within the clades. The tree was inferred by maximum-likelihood analysis of alignments of predicted amino acid sequences of selected ceramide synthases from the ascomycete classes Dothideomycetes, Eurotiomycetes, Leotiomycetes, Pezizomycetes, Saccharomycetes and Sordariomycetes. Sequences of five basidiomycete species were also included in the analysis, and the tree was rooted with rat and human sequences. Accession numbers for protein sequences are indicated after species names. Joint Genome Institute accession numbers are preceded by JGI; all other accessions are from NCBI/GenBank. In the Fum17 and Fum18 clades, the five-digit numbers after some of the Fusarium species names are NRRL strain designation. The genome sequences of these are present in GenBank, but they are not annotated. Numbers near branches are bootstrap values based on 1,000 pseudoreplicates. Values of <70 are not considered to be significant and therefore are not shown.