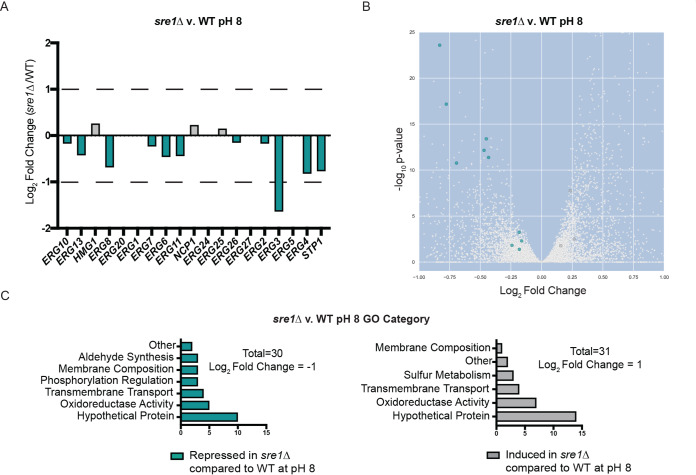
FIG 5.
Transcriptomic analysis of the sre1Δ and wild-type strains in response to alkaline pH. WT and sre1Δ cells were incubated in YPD medium pH 4 or pH 8 for 90 min. This experiment was conducted with six biological replicates for each strain and condition. Total RNA was extracted, mRNA was isolated, and libraries were prepared and finally sequenced using an Illumina NextSeq 500 sequencer. GO-term analysis was performed using FungiDB. (A) The majority of the known genes in C. neoformans ergosterol biosynthesis were significantly differentially expressed in the sre1Δ versus wild-type transcriptome at pH 8. ERG genes that were significantly differentially expressed have an adjusted P value of <0.016 (teal, repressed in the sre1Δ mutant compared to wild type; gray, induced in the sre1Δ mutant compared to wild type). (B) Volcano plot displaying the significantly regulated transcripts in the sre1Δ versus wild-type transcriptome at pH 8 (adjusted P value of <0.05) (teal, repressed in the sre1Δ mutant compared to wild type; gray, induced in the sre1Δ mutant compared to wild type). The full volcano plot (zoomed out) is shown in Fig. S2. (C) GO-term analysis of the sre1Δ versus wild-type differentially expressed genes following a 90-min shift from YPD pH 4 to YPD pH 8. These transcripts were selected based on a strict cutoff of log2 fold change of ±1. Teal, biological processes repressed in sre1Δ mutant compared to wild type at high pH; gray, biological processes induced in sre1Δ mutant compared to wild type.