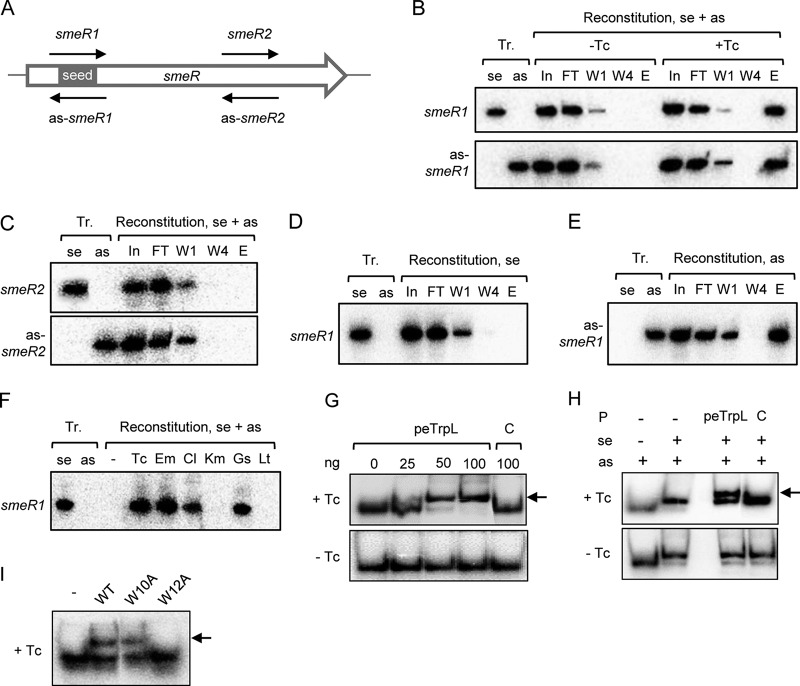
FIG 5.
Reconstitution of ARNP complexes. (A) Scheme of the smeR ORF (white arrow) showing the proposed peTrpL binding seed region and the in vitro transcripts used for ARNP reconstitution (thin black arrows). (B to F) Northern blot analyses with probes detecting the in vitro transcripts indicated on the left side of each panel. At the top of each panel, the loaded samples are indicated. Tr., in vitro transcripts were loaded as hybridization and size controls; se, sense transcript (corresponds to smeR mRNA); as, antisense transcript (corresponds to as-smeR RNA); reconstitution se + as, both sense and antisense transcript were present in the reconstitution reaction; reconstitution se, only the sense transcript was used for reconstitution; reconstitution as, only the antisense transcript was used for reconstitution. The 50-μl reconstitution samples were used for CoIP and following fractions were loaded on the gel (volume or amount loaded): In, input fraction (5 μl); FT, flowthrough (5 μl); W1 and W4, first and last washing fractions (10 μl each); E, elution fraction, 1/10 of the purified CoIP-RNA. Shown are representative results. (B) ARNP reconstitution using the sense transcript smeR1 and the antisense transcript as-smeR1, which correspond to the putative peTrpL binding site (seed region). Addition of Tc is indicated. Top, hybridization with a probe directed against smeR1. Bottom, rehybridization of the membrane with a probe directed against as-smeR1. (C), Reconstitution with the control transcripts smeR2 and as-smeR2 in the presence of Tc. Top, hybridization with a probe directed against smeR2. Bottom, rehybridization of the membrane with a probe directed against as-smeR2. (D) ARNP reconstitution using only smeR1 in the presence of Tc. (E) ARNP reconstitution using only as-smeR1 in the presence of Tc. (F) ARNP reconstitution using smeR1, as-smeR1, and the indicated antibiotics and flavonoids. Only elution fractions were loaded. −, representative negative control of reconstitution, 2 μl ethanol was added to the reconstitution mixture. (G to I) EMSAs in the presence or absence of Tc (indicated on the left) using radioactively labeled as-smeR1 and 10% PAA gels. Shift caused by peptide is indicated by an arrow. (G) shift of as-smeR1 by increasing peTrpL amounts (indicated in nanograms). C, control, unrelated protein was used. (H) Shift of an RNA duplex (smeR1 and as-smeR1 transcripts) by 50 ng peTrpL. Presence of transcripts and proteins in the loaded samples is indicated at the top. (I) WT peTrpL or peptides (50 ng) with the indicated amino acid exchanges were used in the EMSAs with as-smeR1. −, no protein was present.