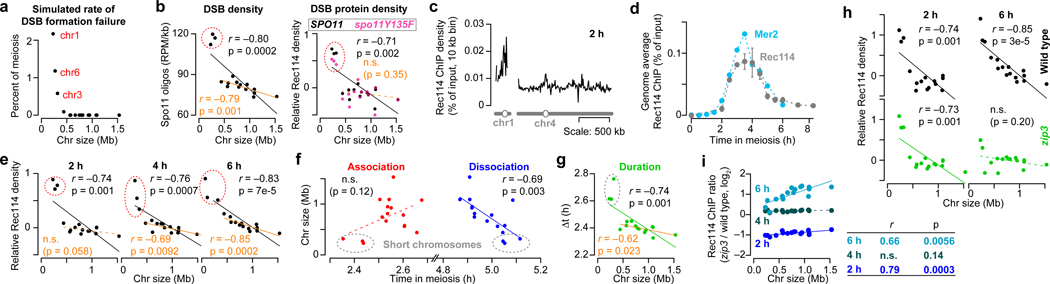
Figure 1. Rec114 and Mer2 accumulate preferentially on smaller chromosomes.
a, Random DSBs. For each simulation, 200 DSBs were distributed randomly by chromosome size; S. cerevisiae makes 150–200 DSBs per meiosis15. The plot shows the percent of simulations (10,000 total) in which the indicated chromosome failed to acquire a DSB.
b, Chromosome size dependence of DSBs (Spo11-oligo density in reads per million (RPM) per kb, 4 h, n = 28 maps), and Rec114 binding (ChIP-chip enrichment, 4 h, n = 1 culture for each strain). In all figures, chr12 is represented without including its full rDNA cluster. Data are from refs3,38. Relative Rec114 density: the mean ChIP signal for each chromosome was normalized to the mean for chr15 and log2-transformed. All correlations reported are Pearson’s r.
c, Example Rec114 ChIP-seq for chr1 and chr4 at 2 h. Absolute ChIP signal was calibrated by qPCR and smoothed with 10-kb sliding window.
d, Time course of genome-average Rec114 and Mer2 levels. Gray points are mean ± range of two Rec114 datasets. Cyan points are from the single Mer2 dataset.
e, Size dependence of per-chromosome Rec114 ChIP density. The full time course is in Extended Data Fig. 1d.
f, Per-chromosome association and dissociation times for Rec114.
g, Per-chromosome Rec114 duration. Panels c and e–g are from the ARS+ time course (n = 1 culture); other time courses are in Extended Data Fig. 1 and 2.
h, i, Zip3-dependent homolog engagement shapes Rec114 abundance late in prophase. Graphs show per-chromosome Rec114 ChIP density (normalized to chr15) (h) and log fold change in the zip3 mutant (i). n = 1 culture for each strain.