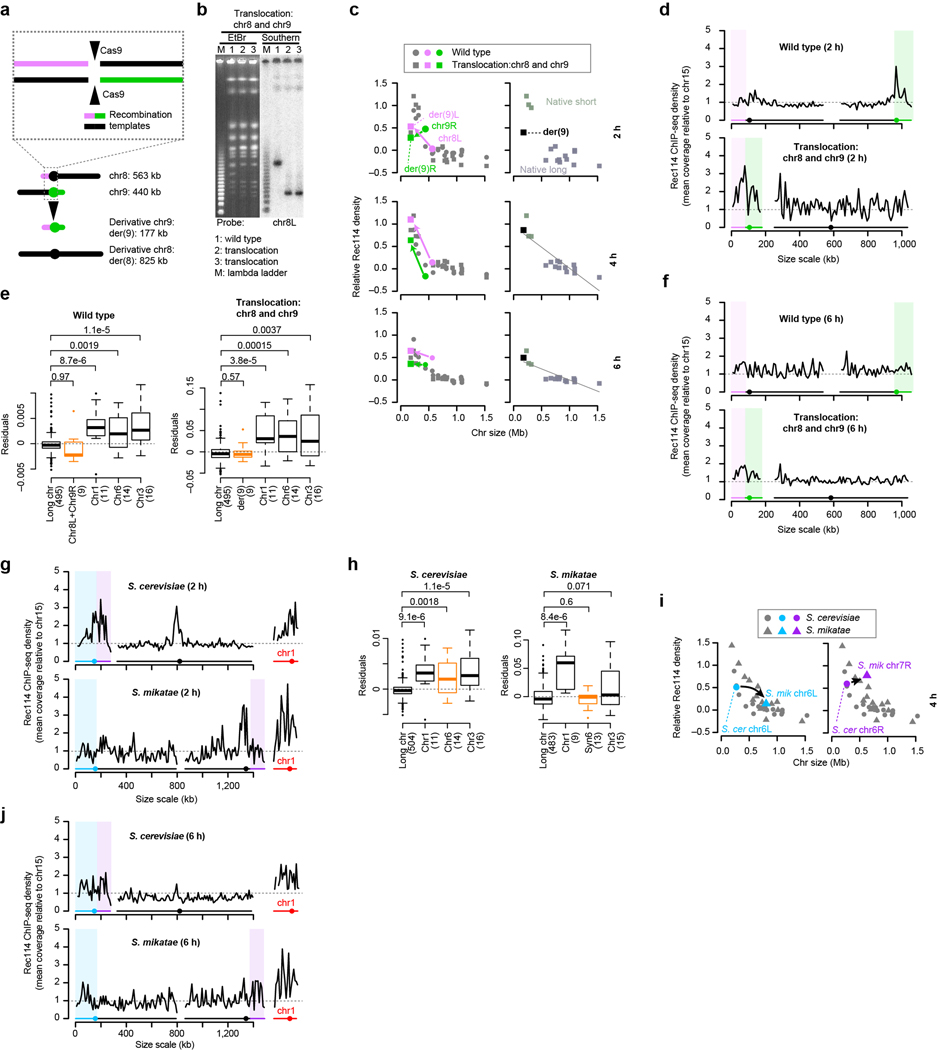
Extended Data Figure 6. An artificially short chromosome fails to acquire a boost in Rec114 binding.
a, Top: Strategy to target reciprocal translocation. To generate the translocation between chr8 and chr9, we introduced a plasmid expressing Cas9 and two guide RNAs that target cleavage within chr8 and chr9, respectively. This plasmid was introduced by co-transforming it along with 100 bp long recombination templates matching the desired reciprocal recombination products. Bottom: Targeted translocation between chr8 and chr9 (to scale).
b, Confirmation of targeted translocations as described in Extended Data Fig. 5f. The translocations were verified by Southern blotting using probes against the right and left ends of both chromosomes involved in the translocation (n = 4 different probes). A representative result using the chr8L probe is presented. For gel source data, see Supplementary Figure 1.
c–f, Per-chromosome Rec114 ChIP densities (c), Rec114 profiles at 2 h and 6 h (d, f), and multiple regression residuals (e) are shown as Fig. 3b, c, and Extended Data Fig. 5g–i. In e, numbers in parentheses indicate number of bins. P values are from one-sided Wilcoxon tests. Boxplots are as described in Extended Data Fig. 2c.
g–j, High-level Rec114 binding to chr6-derived sequences is not retained in S. mikatae. Rec114 profiles (g, j), multiple regression residuals (h) and Rec114 ChIP density at 4 h (i) are shown as in Fig. 3b, c, and Extended Data Fig. 5g–i. In h, numbers in parentheses indicate number of bins. P values are from one-sided Wilcoxon tests. Note that the model also tended to underperform for chr3 in S. mikatae, but the distribution of residuals was not statistically significant (p = 0.071).