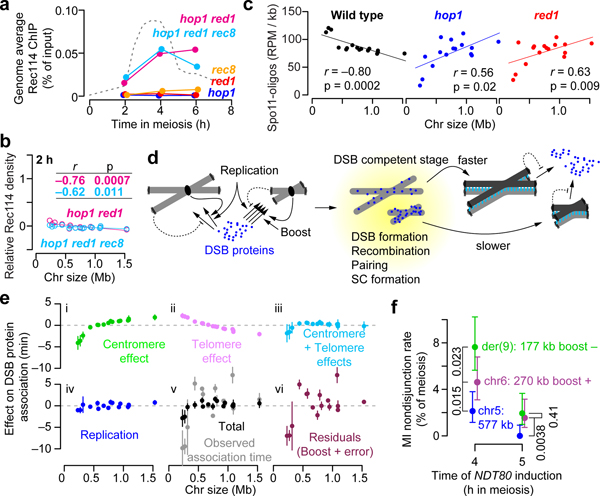
Figure 4. Role of axis proteins in the short chromosome boost and an integrated view of DSB control.
a, Genome-average Rec114 levels. Dashed line, wild-type profile from Fig. 1d.
b, Chromosome size dependence of Rec114 binding requires Hop1 and Red1. Although correlations with size are statistically significant, slopes are negligible compared to wild type (Fig. 1e).
c, Preferential DSB formation on small chromosomes is lost in hop1 and red1 mutants. Samples were collected at 4 h in meiosis. n = 28 (wild type), 2 (hop1 and red1) independent maps.
d, Cartoon depicting the multiple pathways that govern DSB protein association and dissociation from long vs. short chromosomes.
e, Size dependence of each early pathway and their integration. Net effects for each chromosome (relative to mean association time) were estimated from the three-factor models in Fig. 2d and Extended Data Fig. 5a (points and vertical bars are means ± SD of the three data sets). Panels i–iii show centromere and telomere effects and their combination. Panel iv shows the effect of replication timing (high SD for chr3 is from arsΔ). Panel v merges all three pathways and compares this total to observed association time. Panel vi depicts what is not explained by these three pathways, inferred as the short chromosome boost. See Supplementary Discussion 4 for further details.
f, Small chromosomes are at greater risk of missegregation when DSB control is dysregulated. Chromosome missegregation was scored using spore-autonomous fluorescent reporters39 (Extended Data Fig. 10d, e) in a strain with NDT80 under inducible control. Premature NDT80 expression (4 h, n = 563 tetrads) caused more missegregation for smaller chromosomes, especially for der(9) which lacks the small-chromosome boost. Segregation became more accurate when NDT80 was induced later (5 h, n = 516 tetrads) to allow more time for DSBs to continue to accumulate; because of feedback tied to homolog engagement, longer time is predicted to provide greater benefit to smaller chromosomes, as was seen. Error bars represent 95% confidence intervals. P values from one-sided Fisher’s exact test are shown.