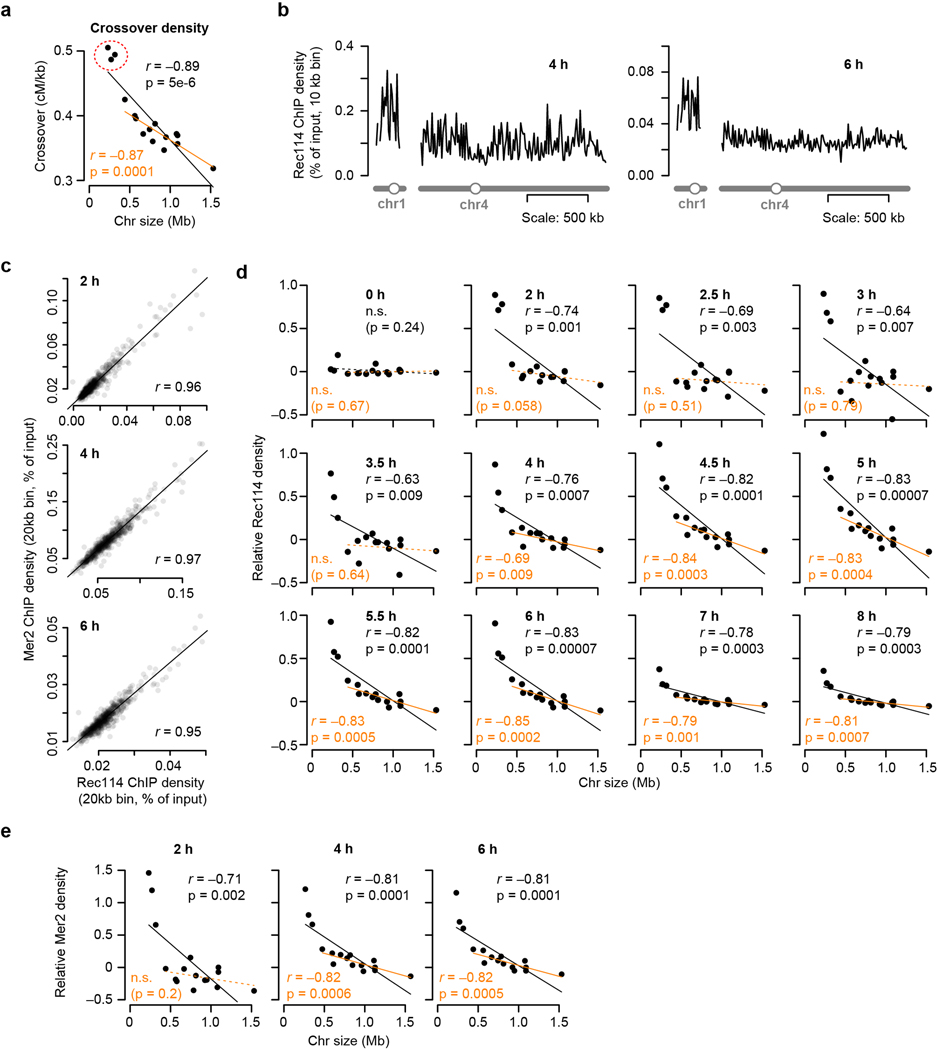
Extended Data Figure 1. Chromosome size dependence of DSB protein binding.
a, Chromosome size dependence of crossovers (centiMorgans (cM) per kb). Data are from ref 12. n = 51 tetrads.
b, Example Rec114 ChIP-seq profiles for chr1 and chr4 at 4 h and 6 h.
c, Similarity of Rec114 and Mer2 ChIP-seq patterns. Both strains are arsΔ background, where all active replication origins on the left arm of chr3 are deleted. Similar profiles of both proteins suggest similar regulation. n = 1 time course for each strain.
d, Full time course of average per-chromosome Rec114 ChIP densities (ARS+ strain). n = 1 time course.
e, The size dependence of average per-chromosome Mer2 ChIP density changes over time similar to Rec114 (compare to Fig. 1e). n = 1 time course.