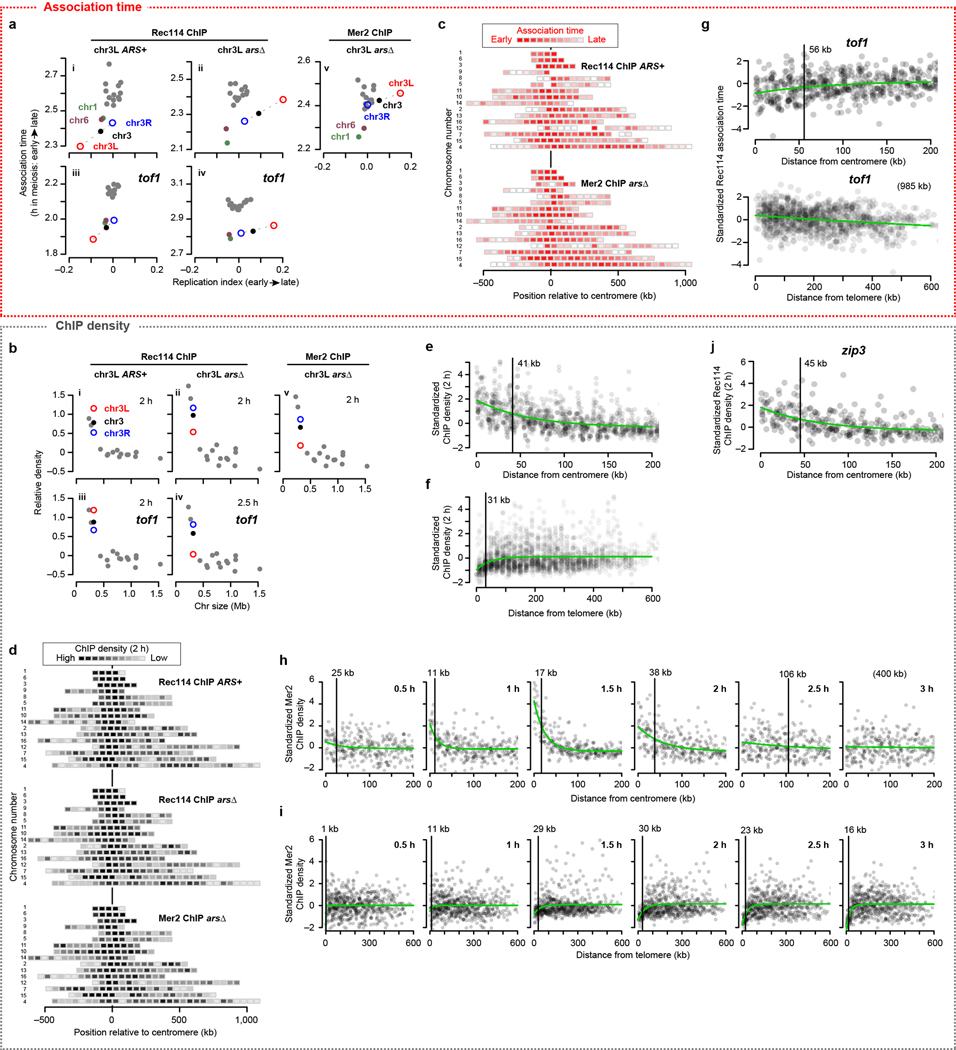
Extended Data Figure 4. Replication timing and the centromere and telomere effects.
a, b, Comparison of per-chromosome Rec114 and Mer2 association time (a) and ChIP density at early time points (b, 2 and 2.5 h, normalized to chr15) with replication timing. Replication index is defined as –log2 of the ratio of sequence coverages for ChIP input samples in S phase vs. G1 phase; lower values indicate earlier replication18. The means for the left arm (chr3L, open red circle) and right arm (chr3R, open blue circle) of chr3 are plotted separately, connected to the means for the entire chromosome (solid black) by dashed lines. In wild type, chr3L showed both early replication and early Rec114 association (a.i). Origin deletion delayed replication and Rec114 association, but association was still on the early side compared with longer chromosomes despite extreme replication delay (a.ii). Mer2 was similar (a.v). Moreover, in tof1, the short trio still showed early Rec114 association (a.iii) and origin deletion delayed Rec114 association18 less than it delayed replication (a.iv). ChIP densities exhibited a complementary trend: Rec114 and Mer2 overrepresentation on short chromosomes was partially dependent on early replication (b). Rec114 was naturally overrepresented on chr3L (b.i). Origin deletion caused a substantial decrease on chr3L but still left Rec114 at higher levels than on the larger chromosomes (b.ii) and Mer2 showed a similar pattern (b.v). In the tof1 mutant, the short trio still showed high Rec114 signal (b.iii), and origin deletion reduced this signal on chr3L but left it in the higher part of the longer-chromosome range (b.iv). The similarity with Rec114 patterns, including much later Mer2 accumulation on the late-replicating left arm of chr3, suggests that Mer2 binding to chromatin is also coordinated with replication timing. Mer2 is able to bind chromatin in the absence of Rec1143,6,40, but interaction with Rec114 (which is promoted by replication-associated Rec114 phosphorylation18) might stabilize or otherwise modify the localization of Mer2.
c, d, Intra-chromosomal distributions of Rec114 and Mer2 association times (c) and ChIP density at 2 h (d) in the indicated strains. c, Average values for 50-kb bins are presented as described in the legend to Fig. 2a. d, Each 50-kb bin is color-coded according to the mean ChIP density within the bin.
e, f, Centromere (e) and telomere (f) effects on ChIP density of DSB proteins at 2 h. The two Rec114 and one Mer2 ChIP time points were binned, standardized, and pooled as in Fig. 2b, c.
g, Centromere (top graph) and telomere (bottom graph) effects on Rec114 association time in tof1 mutants. Rec114 ChIP-seq data from ARS+ tof1 and arsΔ tof1 strains were binned, standardized, and pooled as in Fig. 2b, c. The centromere and telomere effects are still apparent in the tof1 mutants, but appear substantially weaker (particularly the telomere effect).
h, i, Detailed time courses of Mer2 binding to chromatin near centromeres (h) and telomeres (i).
j, The zip3 mutation did not affect the centromere effect on Rec114 ChIP density at 2 h.