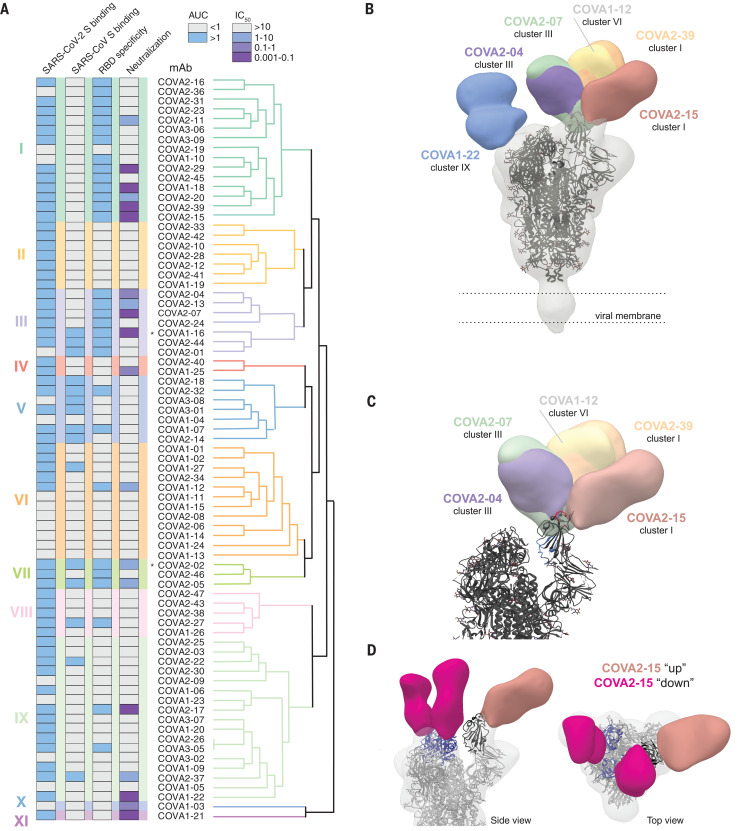
Fig. 5. Antigenic clustering of SARS-CoV-2 S protein–specific mAbs.
(A) Dendrogram showing hierarchical clustering of the SPR-based cross-competition heat map (table S2). Clusters are numbered I to XI and are depicted with color shading. ELISA binding to SARS-CoV-2 S protein, SARS-CoV S protein, and SARS-CoV-2 RBD as presented by AUC and neutralization IC50 (μg/ml) of SARS-CoV-2 is shown in the columns on the left. ELISA AUCs are shown in gray (AUC < 1) or blue (AUC > 1), and neutralization IC50 is shown in gray (>10 μg/ml), blue (1 to 10 μg/ml), violet (0.1 to 1 μg/ml), or purple (0.001 to 0.1 μg/ml). Asterisks indicate antibodies that cross-neutralize SARS-CoV pseudovirus. (B) Composite figure demonstrating binding of NTD-mAb COVA1-22 (blue) and RBD mAbs COVA2-07 (green), COVA2-39 (orange), COVA1-12 (yellow), COVA2-15 (salmon), and COVA2-04 (purple) to SARS-CoV-2 spike (gray). The spike model (PDB 6VYB) is fit into the density. (C) Magnification of SARS-CoV-2 spike comparing epitopes of RBD mAbs with the ACE2-binding site (red) and the epitope of mAb CR3022 (blue). (D) Side (left) and top (right) views of the 3D reconstruction of COVA2-15 bound to SARS-CoV-2 S protein. COVA2-15 binds to both the down (magenta) and up (salmon) conformations of the RBD. The RBDs are colored blue in the down conformation and black in the up conformation. The angle of approach for COVA2-15 enables this broader recognition of the RBD while also partially overlapping with the ACE2-binding site and therefore blocking receptor engagement.