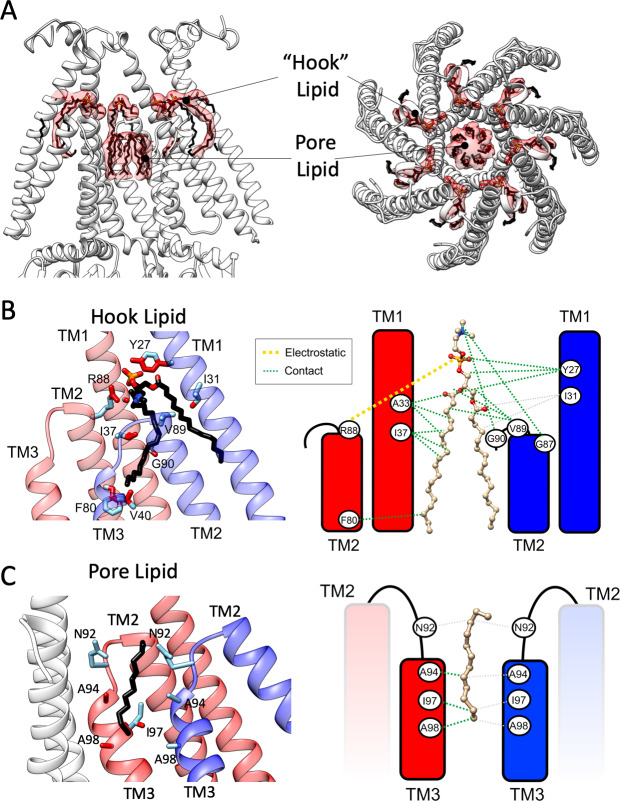
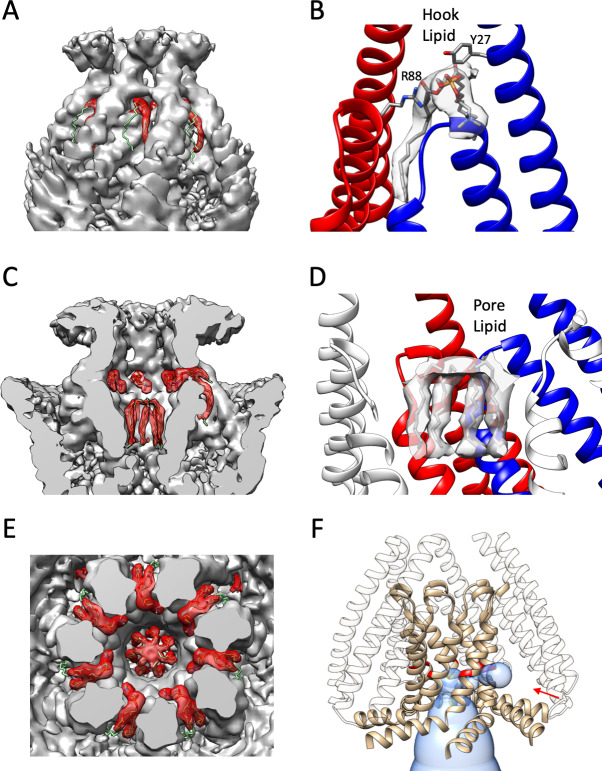
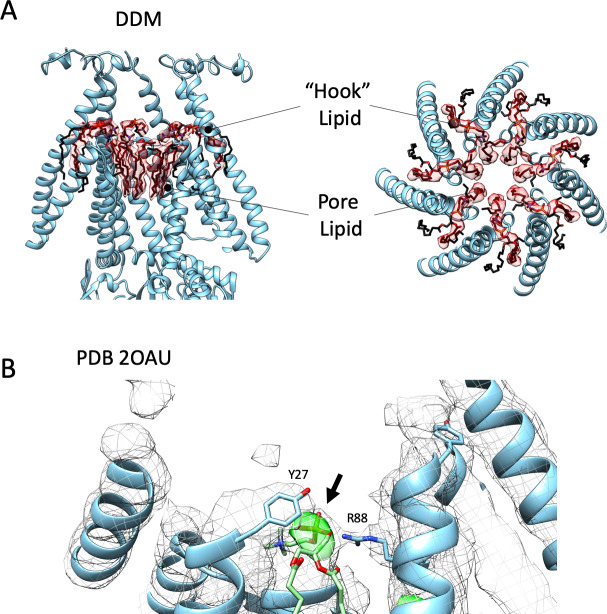
Figure 5. Bound lipid at the inner gate and the permeation pathway.
(A) Side (left) and top (right) views of EM density (transparent red surface) associated with putative lipid molecules bound to MscS-ND (shown in white cartoon representation). A ‘hook’ phospholipid is cradled at the subunit interface atop the TM2-TM3 hairpin, while seven individual acyl chains line the permeation pathway along TM3, above the narrowest portion of the gate. (B) Contact map and coordination of the hook lipid. Left, TM helices from two adjacent subunits (red and blue) are shown. Key interactions are highlighted for R88 (in the red subunit) with the head group nitrogen (PC or PE) and Y27 (in the blue subunit) with the phosphate group. Residues within van der Waals distances are shown in red. Right, a cartoon representation of the contact/coordination map. (C) Same as (B), but with the pore lipid acyl chains.