Figure 2. Reconstitution of l-Opa1.
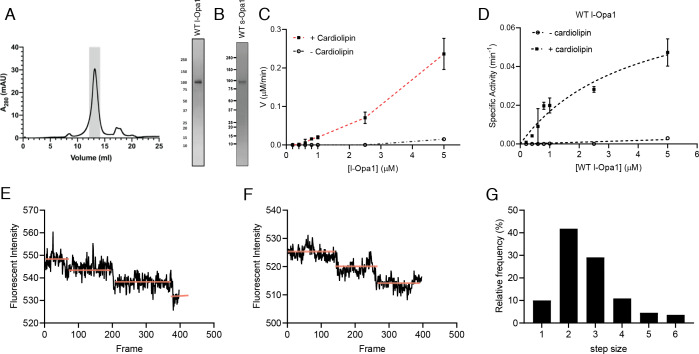
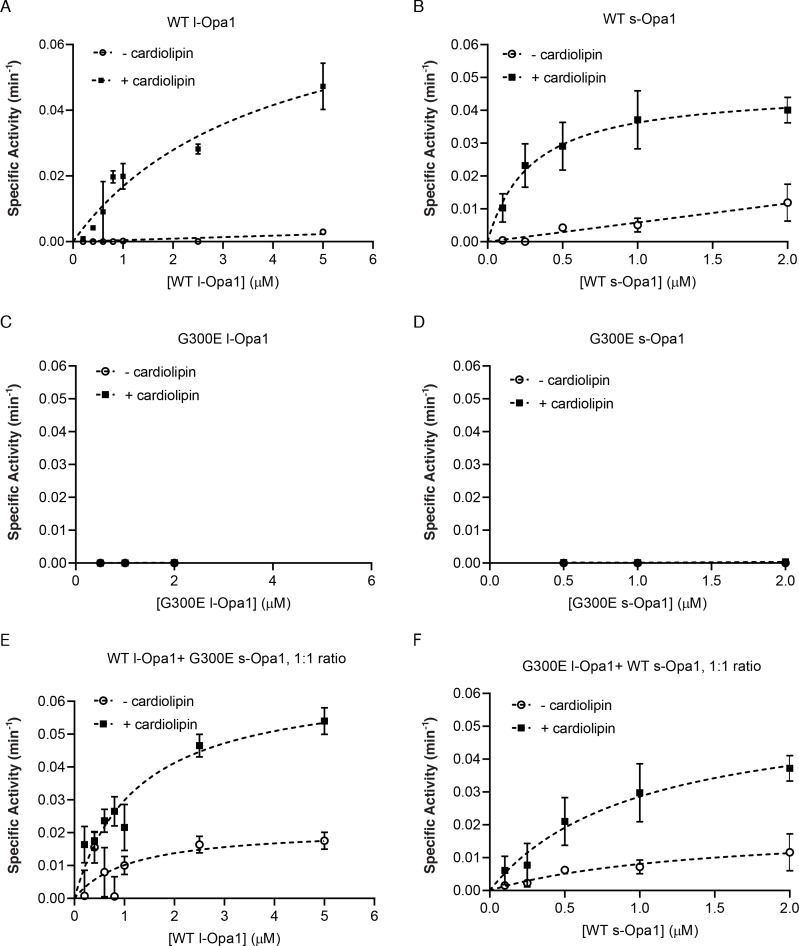
(A) Representative size-exclusion chromatograph and SDS-PAGE gel of human l-Opa1 purified from P. pastoris. (B) SDS-PAGE gel of human s-Opa1 purified from P. pastoris. l-Opa1 activity, with velocity (C) and specific activity (D) of GTP hydrolysis in the presence and absence of cardiolipin, while varying protein concentration of Opa1. Data are shown as mean ± SD, with error bars from three independent experiments. Representative single-liposome photobleaching steps (E and F) and histogram of step sizes (distribution for 110 liposomes shown) (G).
Figure 2—figure supplement 1. GTP hydrolysis activity.
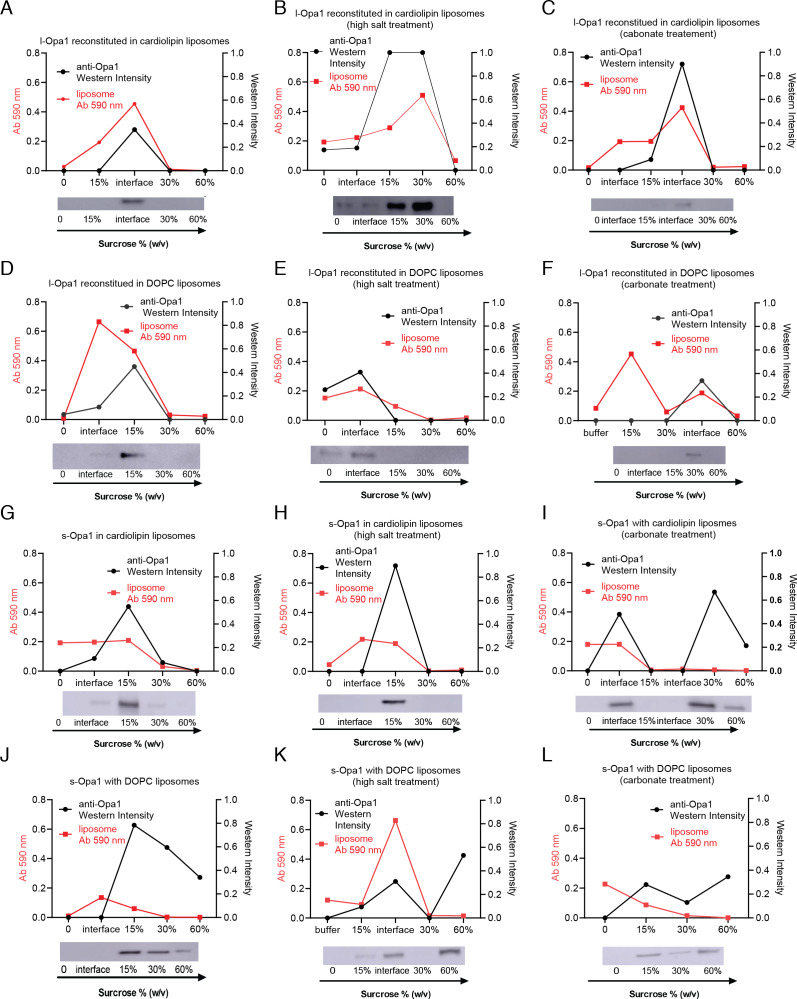
Figure 2—figure supplement 2. Liposome co-flotation.
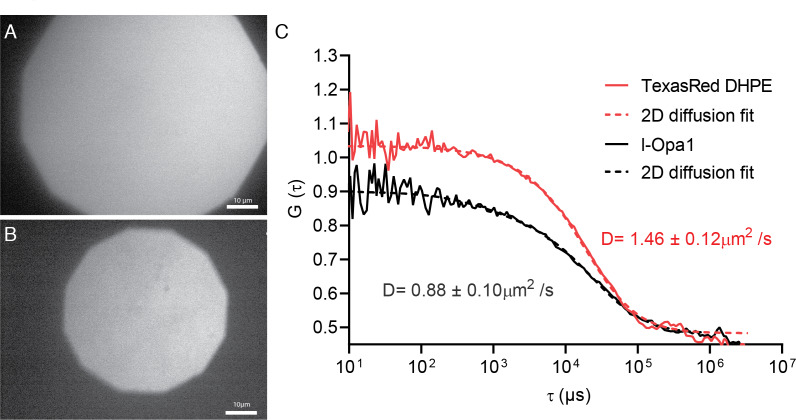
Figure 2—figure supplement 3. Bilayer homogeneity and FCS.
Figure 2—figure supplement 4. Blue native gels.
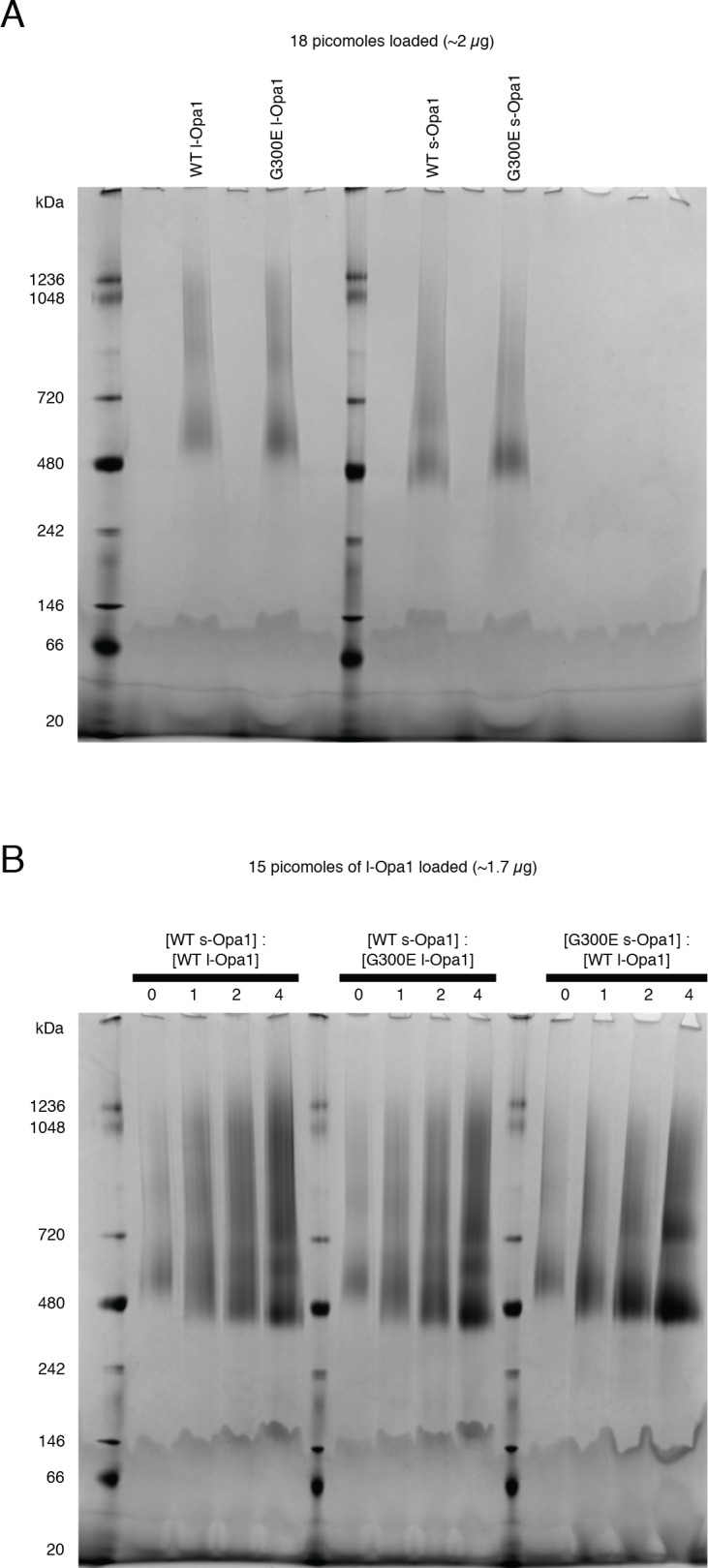
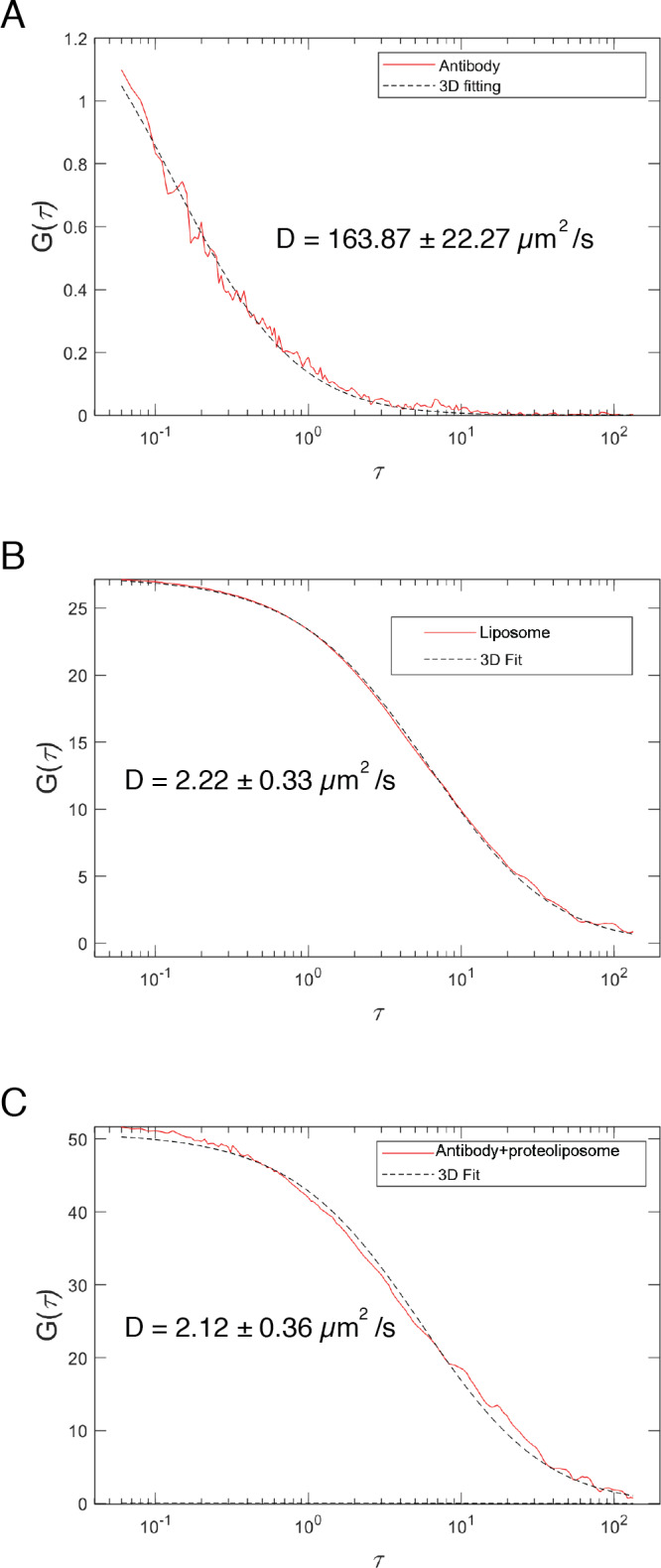
Figure 2—figure supplement 5. FCS.