FIGURE 2.
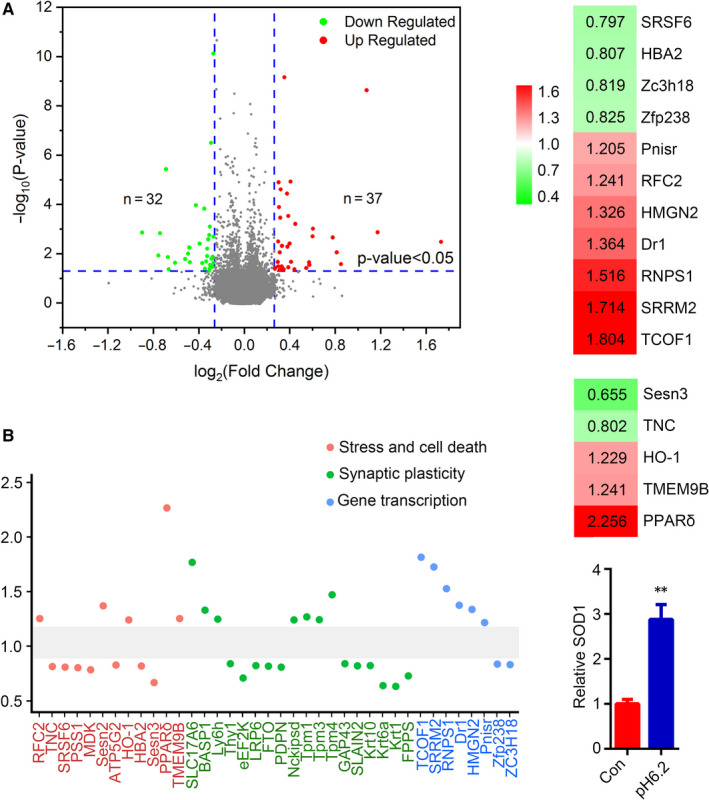
Classification of differentially expressed proteins of pH6.2 neurons. A, A total of 69 differentially expressed proteins were obtained in this study. Among them, there are 37 proteins were up‐regulated (ratio > 1.2, P < 0.05), and 32 proteins were down‐regulated (ratio < 0.83, P < 0.05) in pH6.2/Con. B, By biological functions, the differentially expressed proteins were classified into three groups, for example stress and cell death, synaptic plasticity and gene transcription. C, The differentially expressed proteins related to gene transcription are listed with a ratio (pH6.2/Con), including replication factor C subunit 2 (RFC2), serine/arginine‐rich splicing factor 6 (SRSF6), haemoglobin subunit α 2 (HBA2), treacle ribosome biogenesis factor 1 (TCOF1), serine/arginine repetitive matrix 2 (SRRM2), RNA‐binding protein with serine‐rich domain 1 (RNPS1), down‐regulator of transcription 1 (Dr1), non‐histone chromosomal protein HMG‐17 (HMGN2), Pnisr, zinc finger and BTB domain‐containing protein 18 (Zfp238) and zinc finger CCCH domain‐containing protein 18 (ZC3H18). D, The differentially expressed proteins related to oxidative stress are listed with a ratio (pH6.2/Con), including tenascin C (TNC), haeme oxygenase 1 (HO‐1), sestrin 3 (Sesn3), transmembrane protein 9B (TMEM9B) and peroxisome proliferator–activated receptor delta (PPARδ). Red colour indicates the increased proteins (P < 0.05), and green indicates the decreased ones (P < 0.05). E, Levels of superoxide dismutase 1 (SOD1) in neurons were detected by ELISA. Data were presented as means ± SEM (n = 4/group). **P < 0.01 vs Con
