Figure 1.
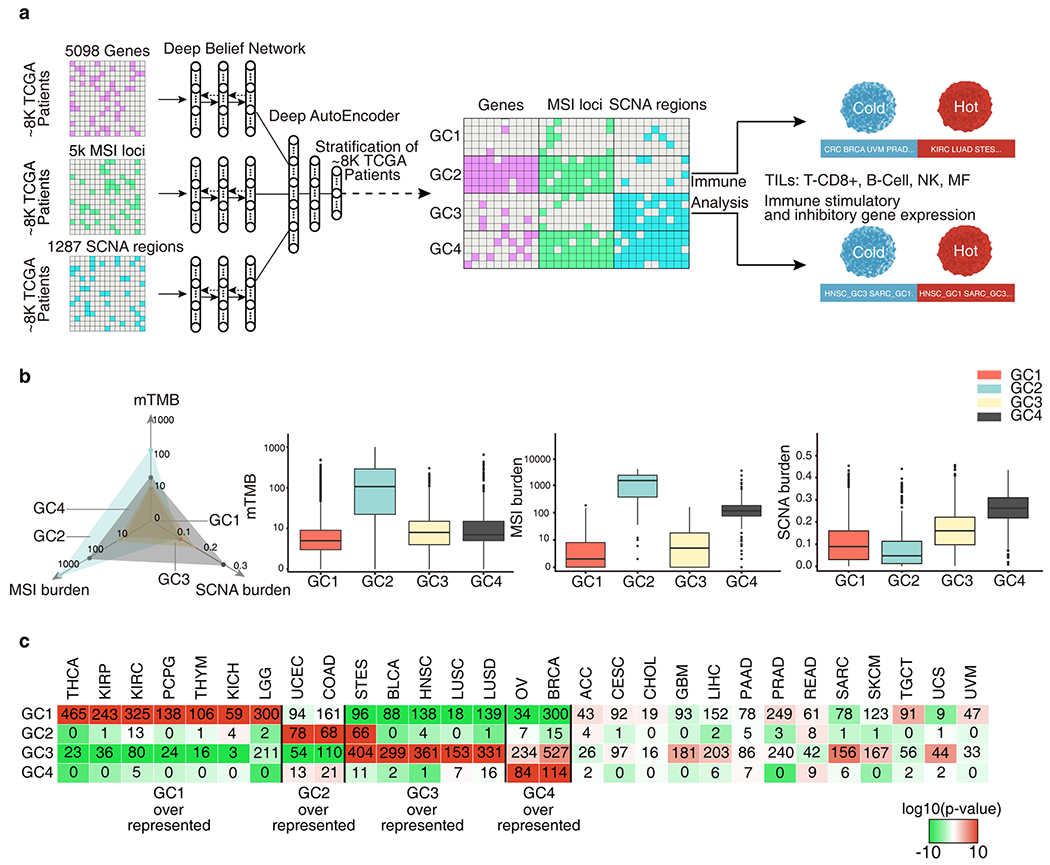
A multifactorial deep learning model reveals novel cancer subtypes and cancer families in terms of intrinsic relationships between the genomic alterations. a, Genomic features of 8,646 TCGA samples across 29 cancer types were extracted from frequently altered somatic mutation genes, microsatellite instability (MSI) loci and somatic copy number alteration (SCNA) regions. A deep learning model was constructed first extracting the high-level presentations of each kind of genomic features using deep belief networks (DBN), then perform deep AutoEncoders (DAEs) on the combined presentations for stratification. Four genomic classes(GCs) were obtained by the stratification. Cancers and cancer subtypes in GCs were identified with immune cold and immune hot microenvironment by the immune gene expression and Tumor-infiltrating lymphocytes (TILs) analysis. b, The mTMB, MSI burden, SCNA burden of each GC. The axis of TMB and MSI burden were scaled by a factor of log10. c, Number of cases in each GC across tumor types. Colors in boxes represent the log10(p-value) calculated from a hypergeometric test comparing the fraction of samples of a given cancer type in a GC to the fraction of samples that are in that GC overall. The red color indicate levels of enrichment while the green color and negative numbers indicate levels of depletion. Samples exceeding an enrichment threshold of 10 are grouped together.
