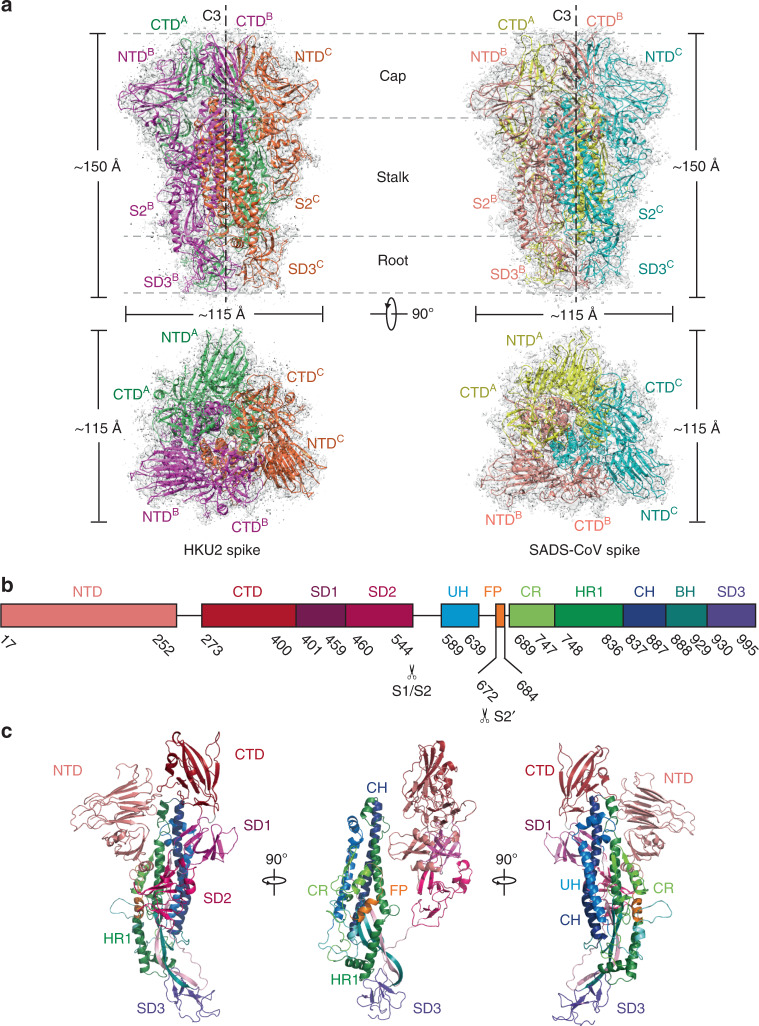
Fig. 1. Overall structures of HKU2 and SADS-CoV spike glycoproteins.
a Overall structures of HKU2 and SADS-CoV spike glycoproteins shown in side view (upper panel) and top view (lower panel). Three monomers of HKU2 spike are colored magenta, green, and orange, respectively; three monomers of SADS-CoV spike are colored pink, yellow, and cyan, respectively. The cryo-EM maps are shown as semitransparent surface and contoured at 2.6 RMS and 3 RMS for HKU2 and SADS-CoV spikes, respectively. The trigonal axes are shown as black dashed lines. Visible segments of each monomer are labeled accordingly. The cap, stalk, and root parts are partitioned by gray dashed lines. b Segmentation of HKU2 monomer. The segments of HKU2 are shown as boxes with the width related to the length of amino acid sequence. The start and end amino acids of each segment are labeled. The position of S1/S2, and S2′ cleavage sites are indicated. NTD N-terminal domain, CTD C-terminal domain, SD1 subdomain 1, SD2 subdomain 2, UH upstream helix, FP fusion peptide, CR connecting region, HR1 heptad repeat 1, CH central helix, BH β-hairpin, SD3 subdomain 3. c Overall structure of HKU2 monomer. Side views of HKU2 monomer shown in three directions. The segments are colored the same as in b.