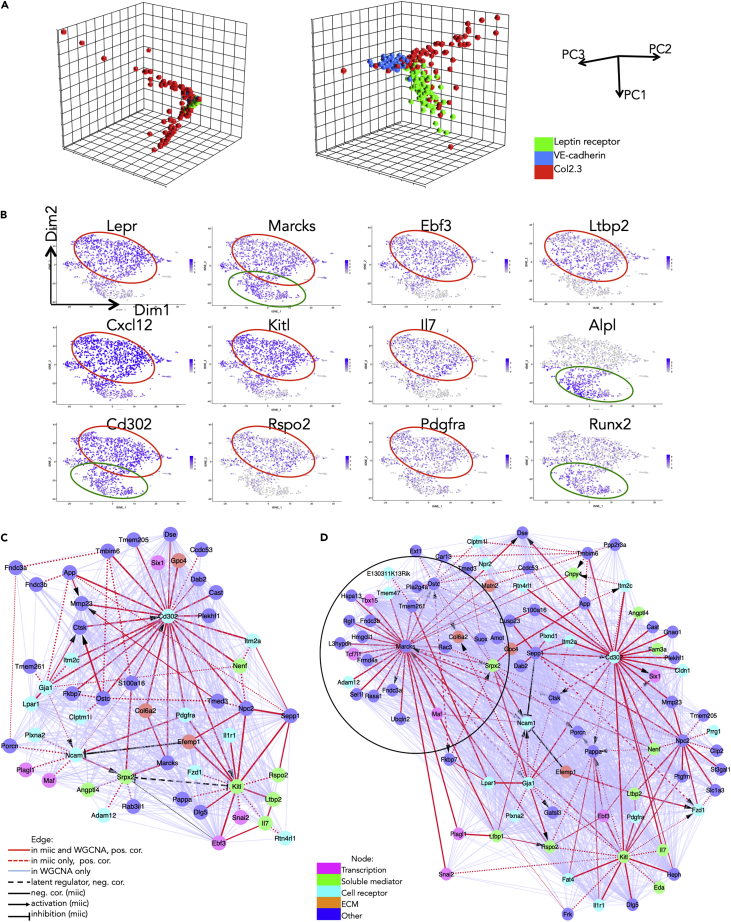
Figure 4.
Single-Cell-Level Analysis Using a Recently Described Algorithm Identifies High-Confidence Paths in the Perivascular Cell Network
(A) PCA using either the whole transcriptome (left) or the Set 3 gene set (right) and 100 randomly selected genes from each of the three populations (VE-cadherin-positive, Leptin receptor-positive, and Col2.3-positive).
(B) t-SNE of example genes expressed in the Leptin receptor-positive population at baseline. All 1,712 cells were included. Negative cells are in gray, and scaled positivity for a given gene is in different hues of blue.
(C) Miic plot of the Set3 genes expressed in the Leptin receptor-positive cell population at baseline. The matrix consisting of 1,712 observations (cells of the Leptin receptor-positive population at baseline) and 109 variables (the Set 3 genes) was submitted to the miic algorithm. 52 genes (nodes) were retained. Edge width increases with degree of confidence in the link as given by miic. The edge line is continuous if the link was found both by miic and WGCNA, dotted if the link was found only by miic, and dashed if there is a common latent regulator found by miic. The edge color in red corresponds to a positive correlation by miic; in black, to a negative correlation by miic; and in light blue, to a positive correlation by WGCNA. The edge head pointed corresponds to an activation, and the edge head blunted corresponds to an inhibition as indicated by miic.
(D) Miic plot of the Set3 genes expressed in the Leptin receptor-positive cell population after stress (treatment with 5-fluorouracil [5-FU]). The matrix consisted of 3,467 observations (cells of the Leptin receptor-positive population after 5-FU treatment) and 109 variables. The new hub and linked targets appearing in condition of stress is encircled.