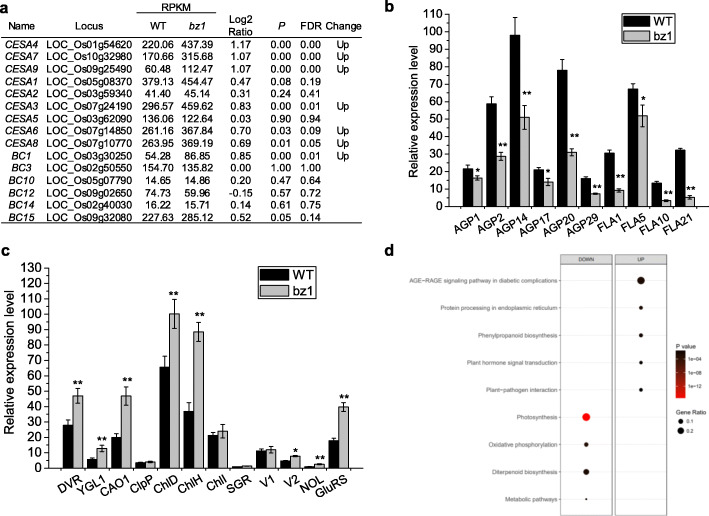
Fig. 7.
Transcriptome profile analysis of both stem and leaf tissues in wild-type (WT) and bz1 by RNA-sequencing approach. a Expression levels of CESA (Cellulose Synthase) and BC (Brittle Culm) genes in WT and bz1 plants based on stem RNA-sequencing analysis. (b) qRT-PCR analysis of AGPs biosynthesis related genes in second stem internodes of WT and bz1 plants at the heading stage. c qRT-PCR analysis of representative genes involved in chloroplast development and chlorophyll biosynthesis using the top leaves of WT and bz1 plants at the tillering stage. d KEGG enrichment analysis was performed to identify the altered pathways of differentially expressed genes (DEGs) based on leaves RNA-sequencing analysis. * and ** indicates significant differences between WT and bz1 by t-test at P < 0.05 and 0.01, respectively