Figure 3.
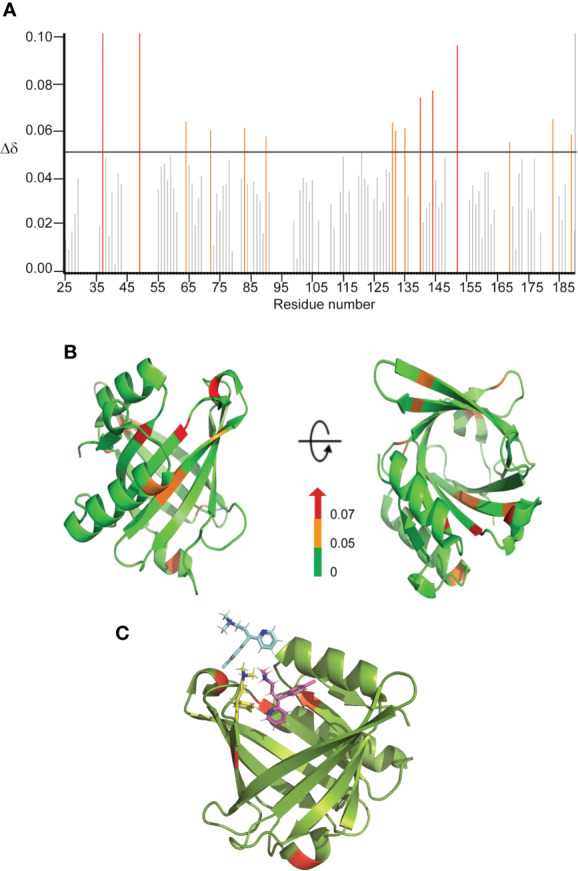
Binding sites characterization of CPM on L-PGDS, as determined by NMR titrations. (A) The chemical shift perturbations (Δδ) of backbone and amide groups of residues in L-PGDS induced by binding to CPM at a molar ratio of 1:4 (protein:drug). (B) Mapping of chemical shift perturbations (Δδ) on L-PGDS crystal structure (PDB code: 4IMN) in the presence of CPM (1:4) (protein:ligand). Red and orange show Δδ >0.07 and 0.05 < Δδ < 0.07, respectively. In the panels, left and right images are the front and top views of L-PGDS (PDB code: 4IMN) in solution, respectively. (C) Model of three CPM molecules (blue, yellow and magenta) docking onto L-PGDS (green) obtained from online protein-ligand docking platform, HADDOCK (van Zundert et al., 2016).
