Figure 3.
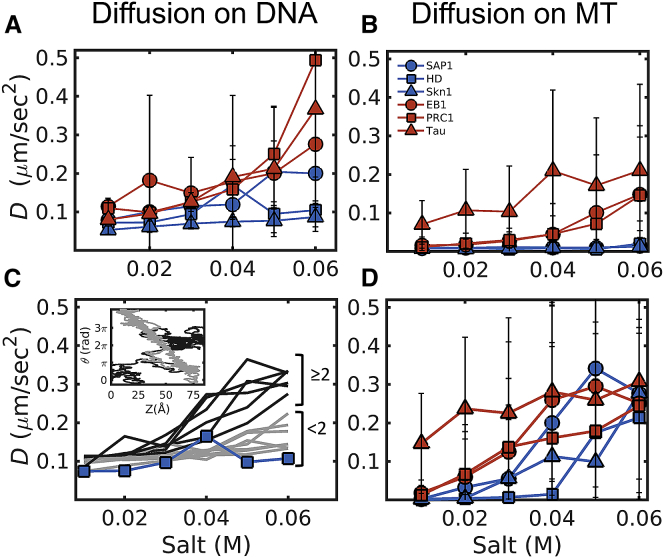
Properties of protein diffusion on DNA and MTs. (A and B) The diffusion coefficients, D, are given as a function of the salt concentration for the three selected DBPs (blue) and three selected MBPs (red) on DNA (A) and on MTs (B). The error bars represent the SD obtained from averaging the independent simulations (see Methods for details). D increases with the salt concentration for the diffusion of both types of proteins on both DNA and MTs. In addition, MBPs diffuse faster than DBPs on both DNA and MTs. (C) This is the same as (A) but for the mutants of DBP HD. Each line represents an HD mutant, in which three positive residues were replaced with three negative residues. The gray lines show mutants with <2 mutations in the recognition helix, and the black lines show mutants with ≥2 mutations in the recognition helix (residues 40–56 of HD). Inset: the coupling between the rotation (θ) around and translation (Z) along the DNA is shown for one HD mutant with <2 mutations in the recognition helix (gray) and one mutant with ≥2 mutations in the recognition helix (black). All the mutants have a positive charge per residue of 0.19 and a net charge per residue of 0.01. (D) This is the same as (B) but for the diffusion on MTs lacking C-terminal tails. Note that at low salt concentrations, MBPs (red) diffuse faster than DBPs (blue) and that this difference diminishes at higher salt concentrations. To see this figure in color, go online.