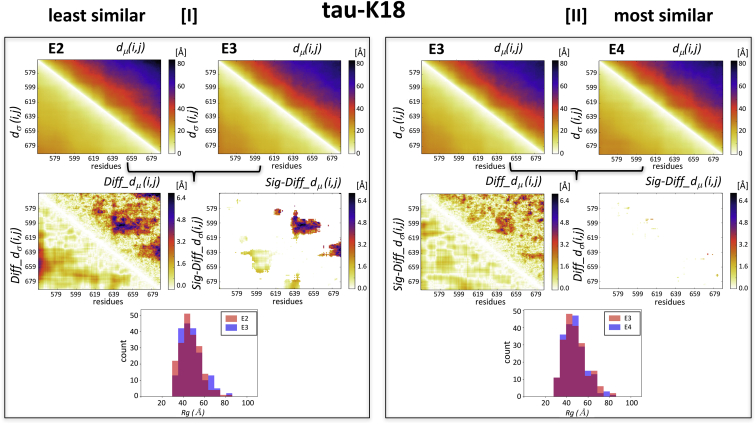
Figure 3.
Comparisons of experimentally characterized tau-K18 IDR ensembles. Illustrated are the similarities between the E2-E3 and E3-E4 pairs of tau-K18 ensembles, displaying the largest (2.15 Å) and smallest (1.47 Å) ens_dRMS-values in Table 1, respectively. (I) Top: shown are the heatmaps of dμ(i,j) and dσ(i,j) matrices for the individual E2 and E3 ensembles. Middle left: shown are the heatmaps of the Diff_dμ(i,j) and Diff_dσ(i,j) computed for the E2-E3 pairs, featuring four regions with the largest differences (>4.8 Å). Middle right: shown are the heatmaps depicting only the statistically significant elements of these maps (Sig-Diff_dμ(i,j) and Sig-Diff_dσ(i,j)). These elements span three regions (residues 578–582 and 619–622, 585–600 and 630–660, and 620–640 and 660–680), representing distances between segments with medium range separation (30–40 residues) along the polypeptide. Bottom: shown is a histogram of the distributions of the gyration radii (Rg) of E2 and E3, which were found to be statistically indistinguishable (p = 0.3). (II) Results for E3-E4 pair are displayed. The top, middle, and bottom panels display the same quantities as in (I), computed for this most similar pair. The Diff_dμ(i,j) and Diff_dσ(i,j) matrices computed for this pair highlight similar differences to those of the E2-E3 pair, but these differences are not statistically significant, resulting in the virtually empty Sig-Diff_dμ(i,j) and Sig-Diff_dσ(i,j) heatmap. The Rg distributions of the E3-E4 pair (bottom plot) are likewise statistically indistinguishable (p = 0.9).