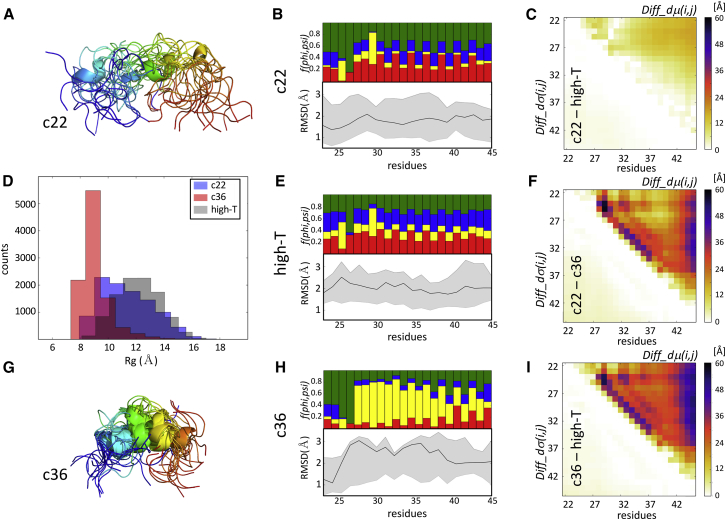
Figure 5.
Comparing ensembles of an intrinsically disordered peptide generated using molecular simulations. Shown is a pictorial summary of the comparative analysis of two ensembles generated using de novo molecular dynamics (MD) simulations for the 24-residue serine-arginine (SR)-rich peptide (residues 22–45 of SR-rich splicing factor 1). These ensembles were generated with the CHARMM22∗ and CHARMM36 force fields, previously reported as featuring the best fit and a poor fit to the experimental data in the original study, respectively (26). (A) Shown is a cartoon representation of the superimposed first 15 conformations from the SR-rich peptide ensemble generated using the CHARMM22∗ force field, which is color coded from blue (N-terminus) to red (C-terminus). (B) Given are twin plots highlighting the regions of the peptide with different backbone flexibility and local-structure preferences in conformations of the CHARMM22∗ ensemble. Bottom graph: medians and 95 percentile confidence interval of the local backbone RMSD distributions for conformations of the ensemble are given. Top graph: fractions of the conformations are given with specific (ϕ,ψ) torsion angle preferences (see the legend of Fig. 1C for details). (C) Shown is a heatmap displaying the statistically significant portions of the Diff_dμ(i,j) and Diff_dσ(i,j) matrices computed between the CHARMM22∗ ensemble and an ensemble generated by the high-temperature MD simulations (High-T) using the same force field. This map shows only small differences, indicating a rather high degree of conformational similarity between the CHARMM22∗ ensemble and its High-T counterpart. (D) Shown is a histogram of the Rg distributions of the ensembles generated using the CHARMM22∗, CHARMM36, and denatured (High-T) ensembles, respectively, illustrating the higher compactness of the CHARMM36 ensemble relative to the other two versions. (E) Shown are twin plots that are analogous to those in (B) computed for the High-T ensemble. (F) Shown is a heatmap displaying the statistically significant portions of the Diff_dμ(i,j) and Diff_dσ(i,j) matrices computed between the CHARMM22∗ and CHARMM36 ensembles. (G) A cartoon representation is given of the superimposed first 15 conformations from the SR-rich peptide ensemble generated using the CHARMM36 force field, which is color coded as in (A). (H) Shown are twin plots highlighting the regions of the peptide with different backbone flexibility and local-structure preferences in conformations of the CHARMM36 ensemble. (I) Shown is a heatmap displaying the statistically significant portions of the Diff_dμ(i,j) and Diff_dσ(i,j) matrices computed between the CHARMM36 and High-T ensembles.