Graphical abstract

Keywords: SARS-CoV-2, Virtual screening, Molecular dynamics, Spike glycoprotein, Sequence alignment, Fusion inhibitors
Highlights
-
•
Up to now, there are no specific vaccines or drugs approved against COVID-19.
-
•
Drug repurposing is a valuable method to identify small-molecule viral inhibitors.
-
•
8770 FDA compounds have been screened using molecular docking and MD simulations.
-
•
A selected set of drugs has been suggested as SARS-CoV-2 fusion inhibitors.
Abstract
The emergence of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has led to a renewed interest in studying the role of the spike S glycoprotein in regulating coronavirus infections in the natural host. Taking advantage of the cryo-electron microscopy structure of SARS-CoV-2 S trimer in the prefusion conformation, we performed a virtual screening simulation with the aim to identify novel molecules that could be used as fusion inhibitors. The spike glycoprotein structure has been completed using modeling techniques and its inner cavity, needful for the postfusion transition of the trimer, has been scanned for the identification of strongly interacting available drugs. Finally, the stability of the protein-drug top complexes has been tested using classical molecular dynamics simulations. The free energy of interaction of the molecules to the spike protein has been evaluated through the MM/GBSA method and per-residue decomposition analysis. Results have been critically discussed considering previous scientific knowledge concerning the selected compounds and sequence alignments have been carried out to evaluate the spike glycoprotein similarity among the betacoronavirus family members. Finally, a cocktail of drugs that may be used as SARS-CoV-2 fusion inhibitors has been suggested.
1. Introduction
The coronavirus disease 2019 (COVID-19) pandemic has rapidly become an international public health threat, which is causing an inconceivable loss of lives and a severe socio-economic instability worldwide. The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), identified as the causal agent of this disease, is an enveloped single-stranded positive-sense RNA virus assigned to the species Severe acute respiratory syndrome-related coronavirus, genus Betacoronavirus, family Coronaviridae, and was firstly identified from a cluster of pneumonia cases in Wuhan (China) on December 2019 (Zhou et al., 2020). On January 30, 2020 the World Health Organization (WHO) declared the coronavirus epidemic as a Public Health Emergency of International Concern. WHO raised the threat of the coronavirus epidemic to the world level to "very high" on February 28, and on March 11 the COVID-19 was officially declared a worldwide pandemic. On March 13, WHO stated that Europe is becoming the new epicentre of the pandemic. The main cause of the uncontainable spread of this disease is the lack of specific antiviral medications, though several groups in the world are making a lot of effort in trying to speed up this research. Although the production of vaccines could be the most powerful weapon against SARS-CoV-2 spread and invasion, it will take quite a long time to develop and clinically test their safety. Thus, as an emergency measure, it would be fundamental to identify or develop antiviral molecules not only to treat the COVID-19 but also to prevent its further transmission.
Although no antiviral treatment has yet been approved, several therapeutic approaches have been proposed, based on the administration of molecules developed for treating other diseases as the combination of lopinavir/ritonavir, chloroquine and hydroxychloroquine (Kupferschmidt and Cohen, 2020). Preclinical studies also suggested that remdesivir, a RNA polymerase inhibitor developed against Ebola, could be effective for both prophylaxis and therapy of human coronavirus infections, as suggested by results obtained testing its activity in a rhesus macaque model of MERS-CoV infection (de Wit et al., 2020). Finally, an investigation led by the Istituto Nazionale Tumori, Fondazione Pascale di Napoli is focused on the use of tocilizumab, a humanized IgG1 monoclonal antibody, directed against the IL-6 receptor and commonly used in the treatment of rheumatoid arthritis, to prevent the cytokine storm syndrome caused by the SARS-CoV-2 infection (ClinicalTrials.gov identifier NCT04317092, available from: https://clinicaltrials.gov/ct2/show/NCT04317092). However, there is no certain evidence that these treatments can be conclusive and that they may also be available for developing countries.
It has been confirmed that the SARS-CoV-2 initiates its entry into host cells by binding to the angiotensin-converting enzyme 2 (ACE2) cell receptor via the receptor binding domain (RBD) of its trimeric spike S glycoprotein, triggering a mechanism which mediates the fusion of host and viral membranes (Zhou et al., 2020). The S protein is a trimeric class I fusion protein that exists in a metastable prefusion conformation, which undergoes substantial structural rearrangements to fuse the viral membrane with the host cell membrane (Bosch et al., 2003; Li, 2016). The S protein further subdivides into two distinct functional subunits: the S1 subunit is involved in host cell receptor recognition, while the S2 mediates viral fusion to host cells. Fusion process is triggered when the S1 subunit binds to a specific cell receptor. Receptor binding destabilizes the prefusion trimer, resulting in shedding of the S1 subunit and transition of the S2 subunit to a stable postfusion conformation. The latter is characterized by the assembly of its heptad repeats 1 (HR1) and 2 (HR2) regions into a 6-helix bundle structure that allows viral fusion and entry into host cells (Bosch et al., 2003; Walls et al., 2017). The complete three-dimensional structure of the human SARS-CoV-2 spike in postfusion conformation has not yet been published but recently Wrapp and coworkers (Wrapp et al., 2020) determined the first 3.5 Å resolution cryo-electron microscopy (cryo-EM) structure of the SARS-CoV-2 S trimer in the prefusion conformation. The study provides biophysical and structural evidences that the SARS-CoV-2 S protein binds the ACE2 receptor with higher affinity than does the closely related severe acute respiratory syndrome coronavirus (SARS-CoV) S protein. Additionally, the authors tested several published SARS-CoV RBD-specific monoclonal antibodies and found no appreciable binding to the SARS-CoV-2 S protein, suggesting that antibody cross-reactivity may be limited between the RBDs of these two viruses (Wrapp et al., 2020).
Because of the crucial function of the S protein in the SARS-CoV-2 infection process, this structural component could represent a target for antibody or small-molecules mediated neutralization, and characterization of the prefusion S structure allowed to get atomic-level information essential to guide the design and development of novel therapeutic molecules.
Previous data obtained for the human respiratory syncytial virus (hRSV) F protein, a class I viral fusion glycoprotein sharing structural similarities with most of the lower respiratory tract viruses’ fusion glycoproteins, indicated that newly generated escape mutations for evaluated small-molecule fusion inhibitors were clustered inside the central cavity of the F protein in prefusion conformation (Battles and McLellan, 2019; Lopes et al., 2020; Yan et al., 2014). Crystallographic studies later confirmed that these small-molecules bind within the central cavity, contacting from two to three lobes of the trimeric binding pocket and interacting with residues of the fusion peptide and the second heptad repeat (Battles et al., 2016). Functional studies involving soluble F proteins (Samuel et al., 2015) and membrane-bound F proteins (Battles et al., 2016), further demonstrated that the evaluated small-molecules act as antagonists that prevent the protein transition to the postfusion conformation. To date, all known small-molecule fusion inhibitors bind in the same pocket and have the same mechanism of action (DeVincenzo et al., 2014; Stevens et al., 2018). These data suggest that, also in the case of SARS-CoV-2, targeting the central cavity of the spike glycoprotein in prefusion conformation could be a valuable approach for hindering its prefusion to postfusion transition and consequently virus entry into host cells.
In this paper, we have used the SARS-CoV-2 S trimer structure in prefusion conformation (Wrapp et al., 2020) to perform a drug repurposing with the aim to find and suggest small-molecule fusion inhibitors for clinical use. Repurposing of drugs already approved, or still in clinical trial, represents an accelerated route for drug discovery, because existing drugs have already established clinical and pharmacokinetic data. We have carried out a virtual screening (VS) simulation using 8.770 FDA drugs extracted from the DrugBank database (https://www.drugbank.ca/) (Wishart et al., 2018). The structure of the spike glycoprotein (Wrapp et al., 2020) has been completed using modeling techniques and used for the identification of available compounds strongly interacting inside its inner cavity (Fig. 1 ). Furthermore, the stability of selected protein-drug complexes has been tested using classical molecular dynamics (MD) simulations and the free energy of interaction of these molecules has been finely evaluated through the molecular mechanics energies combined with generalized Born and surface area continuum solvation (MM/GBSA) method and per-residue energy decomposition analysis (Genheden and Ryde, 2015). Finally, alignments between the sequence of the spike glycoprotein used in our study and sequences of all betacoronavirus spike glycoproteins reported in the GenBank database (Clark et al., 2016), including recently deposited sequences of different SARS-CoV-2 isolates, have been carried out to evaluate the presence of variability within the protein region analyzed by this simulation research.
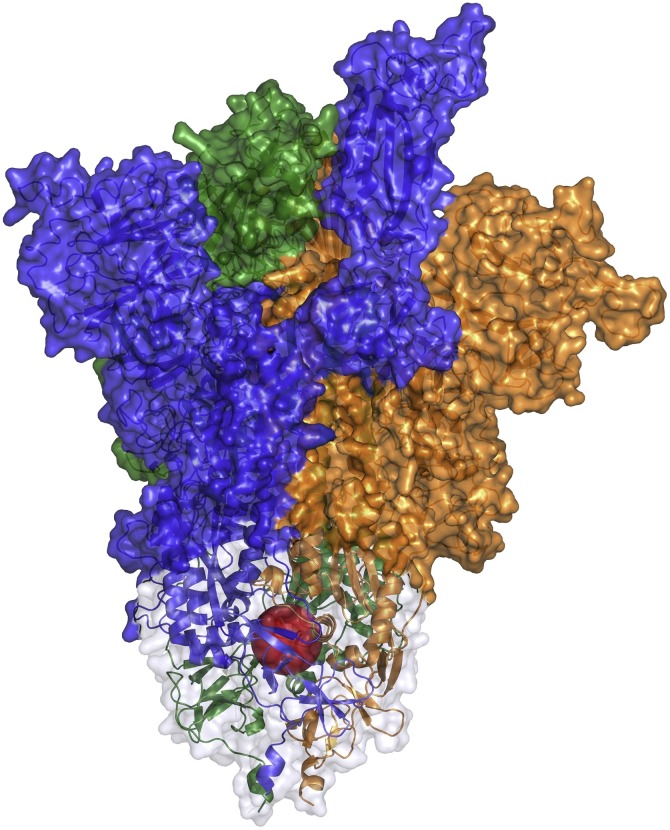
Fig. 1.
Spike S glycoprotein modeled structure (PDB ID: 6vsb). A red sphere has been placed within the structure to highlight the internal cavity, corresponding to the site selected for the Virtual Screening procedure. The HR1 regions of the spike S glycoprotein are shown in cartoon representation surrounded by a light grey transparent surface. The three glycoprotein monomers have been represented with different coloured surfaces (blue: monomer A; orange: monomer B; green: monomer C). This picture has been obtained using the PyMOL 2.1.0 program (The PyMOL Molecular Graphics System, 2020).
As a result, considering the available scientific information, a selected set of drugs has been suggested as SARS-CoV-2 fusion inhibitors, with the hypothesis that the use of these compounds can be extended also to other related viruses.
2. Methods
2.1. Molecular modeling
Cryo-EM structure of the SARS-CoV-2 spike glycoprotein in prefusion conformation has been obtained from the PDB database (PDB ID: 6vsb) (Wrapp et al., 2020). The trimer structure, composed by three identical monomers (named A, B, C), has been completed modeling non-terminal missing loops using the SWISS-MODEL web-server (Waterhouse et al., 2018). The modeling procedure mainly involved regions of the S1 subunit of the glycoprotein (residues: 67–78, 96–98, 143–155, 177–186, 247–260, 329–334, 444–448, 455–490, 501–502, 621–639, 673–686; of monomer A, residues: 67–80, 142–154, 177–186, 210–216, 243–262, 444–448, 455–459, 474–486, 501–502, 621–637, 673–686; of monomer B and residues: 67–79, 96–98, 141–156, 177–186, 246–260, 444–448, 455–459, 472–486, 499–502, 621–640, 673–686; of monomer C), and also two regions of the S2 subunit (residues 812–814 and 829–852 of monomers A, B and C).
Although the modeled regions were not close to the central cavity of the spike glycoprotein, they have been rebuilt since it is not possible to accurately simulate and analyze a protein with missing parts using classical MD simulation. In fact, during the simulation, the protein would be subjected to unnatural fluctuations, misleading the subsequent analyses.
2.2. Virtual screening procedure
A drug library, containing 8.770 compounds in MOL2 format, has been extracted from the DrugBank database v. 5.1.5 (Wishart et al., 2018) (https://www.drugbank.ca/). Spike glycoprotein receptor and drug structure files have been converted into pdbqt format using the prepare_receptor4.py and the prepare_ligand4.py tools of the AutoDockTools4 software (Morris et al., 2009; Sanner, 1999). Seven drug structures have been excluded from the analysis, since prepare_ligand could not generate their PDBQT files, while other eight structures, containing Si atoms, have been excluded due to the absence of force-field parameters. The 8755 molecular docking simulations have been performed using an in-house parallelized version of the Autodock Vina 1.1.2 program (Trott and Olson, 2010). One docking simulation, including ten docking runs, has been carried out for each drug. For each docking simulation, the AutoDock Vina program selects 10 binding poses representing the cluster centroids of all the different conformations, generated in each run using the Lamarckian Genetic Algorithm. A box of size x = 23.25 Å, y = 24.38 Å, z = 25.88 Å has been placed over the HR1 internal region (residues 897–920) of the spike glycoprotein A monomer. 15 receptor residues side-chains around the selected binding site have been considered as flexible (Y904, F906, N907, I909, V911, T912, Q913, N914, V915, D1092, Q1106, N1108 of the monomer A and R1091, E1092, F1121 of the monomer C). Virtual screening has been performed using 3 nodes of the ENEA HPC cluster CRESCO6 (Ponti et al., 2014), where each docking simulation took about 30’. For the 10 top-ranking docked compounds, binding energies have been re-evaluated as an average of the best poses obtained in three repeated molecular docking simulations.
2.3. Molecular docking simulations of multiple compounds
Two sequential molecular docking simulations have been performed for phthalocyanine and hypericin, the two top-ranking drugs, in order to dock a second and a third molecule of the compounds inside the spike glycoprotein binding pocket. The best complexes obtained in the virtual screening first docking run have been used as receptors, converting the structures into pdbqt format using the prepare_receptor4.py tool of the AutoDockTools4 software (Morris et al., 2009; Sanner, 1999). The two molecular docking simulations, each including ten docking runs, have been carried out using the Autodock Vina 1.1.2 program (Trott and Olson, 2010). A box of size x = 25.88 Å, y = 24.00 Å, z = 25.88 Å has been centred over the HR1 internal region of the spike glycoprotein monomer B, selecting 15 residues side-chains around this binding site as flexible (Y904, F906, N907, I909, V911, T912, Q913, N914, V915, E1092, Q1106, N1108 of the monomer B and R1091, E1092, F1121 of the monomer A). Finally, a box of size x = 24.75 Å, y = 25.50 Å, z = 25.88 Å has been placed over the HR1 internal region belonging to the monomer C, selecting 15 side chains as flexible (Y904, F906, N907, I909, V911, T912, Q913, N914, V915, E1092, Q1106, N1108 of the monomer C and R1091, E1092, F1121 of the monomer B). Binding energies have been calculated as an average of the best poses obtained from three repeated docking simulations.
The sequential molecular docking simulations of the 4 top drugs, obtained after re-evaluating the compounds ranking, have been performed for the first three docked compounds applying the same parameters already described for the 3 compounds molecular docking. Molecular docking of the fourth compound, instead, was performed using a box of size x = 27.00 Å, y = 30.38 Å, z = 25.50 Å centred between the HR1 internal regions of the monomers B and C, selecting 12 receptor side chains as flexible (I909, T912, E1092, Q1106, R1107, N1108, F1109 of the monomer B and Y904, R905, N907, Q1036, K1038 of the monomer C).
All molecular docking simulations took about 30’ and have been performed using the ENEA HPC cluster CRESCO6 (Ponti et al., 2014).
2.4. Classical MD simulations of multiple compounds complexes
The two complexes obtained with multiple docking of phthalocyanine and hypericin drugs have been simulated using classical molecular dynamics. Topologies and coordinates files of the input structures have been generated using the tLeap module of the AmberTools 19 package (Case et al., 2018). The spike glycoprotein has been parametrized using the ff19SB force field (Tian et al., 2020), while parameters for the two top-ranking drugs have been generated using the antechamber module of the AmberTools 19 package (Case et al., 2018) and the general Amber force field (Wang et al., 2004). Each spike glycoprotein, complexed with three drugs, has been inserted into a rectangular box of TIP3P water molecules (Jorgensen et al., 1983), setting a minimum distance of 12.0 Å from the box sides and neutralizing the solution with 0.15 mol/L of NaCl ions. In order to remove unfavourable interactions, structures have been subjected to four minimization cycles, each composed by 500 steps of steepest descent minimization followed by 1500 steps of conjugated gradient minimization. A starting restraint of 20.0 kcal*mol−1 *Å-2 has been imposed on protein and ligand atoms and subsequently reduced and removed in the last minimization cycle. Systems were gradually heated from 0 to 300 K in a NVT ensemble over a period of 2.0 ns using the Langevin thermostat (Goga et al., 2012), imposing a starting restraint of 0.5 kcal*mol−1 *Å-2 on each protein and ligand atom, which was decreased every 500 ps in order to slowly relax the system. The systems were simulated in an isobaric-isothermal (NPT) ensemble for 2.0 ns, imposing a pressure of 1.0 atm using the Langevin barostat (Aoki et al., 2004) and fixing the temperature at 300 K. Covalent bonds involving hydrogen atoms have been constrained using the SHAKE algorithm (Ryckaert et al., 1977). A production run of 30 ns has been performed for both systems with a timestep of 2.0 fs, using the pmemd.cuda module of the AMBER 18 software (Case et al., 2018). The PME method was used for calculating long-range interactions while a cut-off of 9.0 Å has been set for short-range interactions (Darden et al., 1993). System coordinates have been saved every 1000 steps.
2.5. Trajectory analysis
RMSD analysis for the spike glycoprotein has been performed using the rms module of the GROMACS 2019 analysis tool (Abraham et al., 2015). Hydrogen bond analyses have been carried out using the HBonds plugin of the software VMD 1.9.3 (Humphrey et al., 1996). Generalized Born and surface area continuum solvation (MM/GBSA) analysis (Genheden and Ryde, 2015) and per-residue energy decomposition analysis have been performed over the last 10 ns of both simulations, using the MMPBSA.py.MPI program implemented in the AMBER18 software (Case et al., 2018) on 3 nodes of the ENEA HPC cluster CRESCO6 (Ponti et al., 2014). Decomposition analysis has been performed on residues surrounding each drug, located within a cut-off distance of 12 Å.
3. Results and discussion
3.1. Virtual screening
A Virtual Screening (VS) analysis (drug repurposing) of a set of 8755 structures of approved, experimental and investigational drugs, extracted from the DrugBank database (https://www.drugbank.ca/) (Wishart et al., 2018), has been performed in order to find compounds that, entering the inner cavity of the protein and blocking the HR1 conformational changes, could act as spike glycoprotein fusion inhibitors.
Using an in-house parallelized version of the AutoDock Vina software (Trott and Olson, 2010), we evaluated the binding affinity of each drug for one of the three HR1 internal regions of the spike glycoprotein (residues 897–920). The ten top-ranking compounds obtained from the VS analysis are reported in Fig. 2 and in Table S1. Interestingly, three of these compounds have already been reported as possible antiviral agents, targeting different types of enveloped viruses. In particular, the compounds 31h-phthalocyanine (from here simply referred to as phthalocyanine) and hypericin represent the two most appealing molecules. In fact, they show a significantly high affinity for the HR1 region and, additionally, hypericin and different phthalocyanine derivates have been already shown to prevent human immunodeficiency virus (HIV) infections, interfering with the surface glycoprotein gp120 binding and fusion to the host cell (François et al., 2009; Lenard et al., 1993; Vzorov et al., 2003). Furthermore, these two compounds, or their derivatives, also possess different antiviral activities towards other viral species, as it will be discussed below in Section 3.5, and hypericin has also been recently shown to inhibit the infectious bronchitis virus (IBV), an avian coronavirus, acting through a mechanism involving inhibition of the virus-induced cellular apoptosis (Chen et al., 2019b).
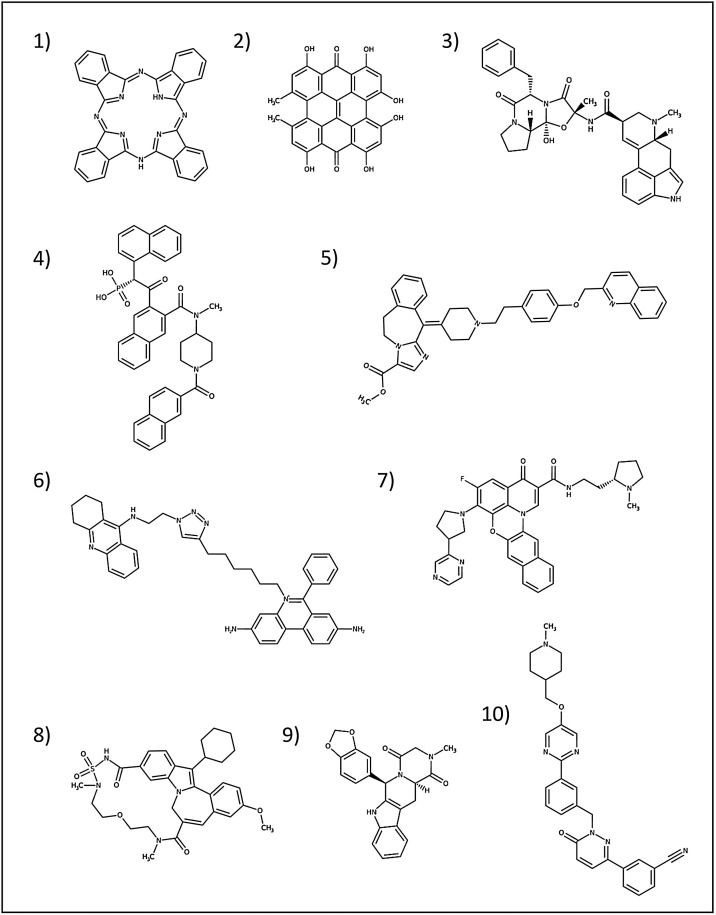
Fig. 2.
Best 10 high affinity compounds identified through the Virtual Screening procedure. 2D representations have been obtained from the DrugBank database (Wishart et al., 2018) (https://www.drugbank.ca/). Drugs order follows that of Table S1.
3.2. Multiple molecular docking simulations within the spike glycoprotein inner cavity
Considering the trimeric structure of the spike glycoprotein, we propose an inhibitory mechanism by which at least three drug molecules could enter and arrange inside the inner cavity, binding the three HR1 internal regions (Wrapp et al., 2020). The concurrent binding of three molecules could generate a strong steric hindrance inside the pocket, preventing the conformational changes of the HR1 regions and hence the formation of the 6-helix bundle stabilizing the postfusion conformation (Walls et al., 2017).
In order to evaluate our hypothesis, starting from the top ranking complexes obtained from the VS procedure, we evaluated the binding affinity of two phthalocyanine or two hypericin molecules for the other two HR1 internal regions of the spike glycoprotein. The average binding energies obtained for each molecule have been reported in Table 1 . It can be observed that even if the binding affinities of the second and third bound molecules are lower than that obtained for the first, due to the reduction in the conformational space inside the pocket, they still remain particularly suitable for establishing valuable interactions. Both compounds contact a high number of residues within the pocket, both from the HR1 internal regions and from upward and downward regions of the spike, creating a connection between the HR1s and surrounding regions of the inner cavity (Fig. 3 ).
Table 1.
Binding energies of three sequential molecular docking simulations performed for phthalocyanine and hypericin. Binding energies are calculated as an average of 3 repeated molecular docking simulations.
| Compound | Binding energy #1 (kcal/mol) | Binding energy #2 (kcal/mol) | Binding energy #3 (kcal/mol) |
|---|---|---|---|
| phthalocyanine | −16.3 | −16.0 | −14.6 |
| hypericin | −15.1 | −14.8 | −12.2 |
Fig. 3.
Best molecular docking complexes of three phthalocyanine (A-C) and three hypericin (D-F) molecules within the inner cavity of the spike glycoprotein. A, D) Drugs are shown as spheres, coloured by atom type with carbon atoms represented in grey. The spike glycoprotein is shown in blue cartoon representation with the HR1 regions highlighted in red. Molecular surface of the protein is shown in transparency. Residues 703-717 and 1070-1138 of the monomer A have been hidden. These images have been generated using the PyMOL 2.1.0 program (The PyMOL Molecular Graphics System, 2020). B, E) 3D representation of the main hydrophobic contacts established by the drugs within the pocket. Drugs and surrounding residues are represented in stick model and coloured by atom type, following the colour scheme described in the previous image. The pictures have been produced using the UCSF Chimera 1.13.1 program (Pettersen et al., 2004). C, F) 2D schematic view of the interactions established by the first docked molecule within the inner cavity of the spike glycoprotein. Hydrogen bonds are shown as green dashed lines between the interacting partners, represented as brown sticks. Hydrophobic interactions are shown as radial half-circles. Drugs bonds and carbon atoms are represented as blue sticks and black filled circles, respectively. Labels indicate residue names in one-letter code, residue numbers and the spike monomer to which each residue belongs. Pictures have been created using the LigPlot+ 1.4 software (Laskowski and Swindells, 2011). PHT: phthalocyanine, HYP: hypericin.
Phthalocyanine molecules arrange in a symmetric orientation (Fig. 3 A–B), facing the inner cavity of the pocket and establishing 11 hydrophobic interactions with surrounding residues. In particular, stacking interactions are settled between the isoindole groups of phthalocyanines and aromatic side-chains of the protein (Fig. 3 B). The three phthalocyanines can also set up hydrophobic interactions within themselves, increasing the stability of the multiple complex (Fig. 3 B–C).
Hypericin molecules, on the other hand, bind to the HR1 regions in an alternate orientation (Fig. 3 D–E), establishing 7 hydrophobic interactions and 4 hydrogen bonds with the surrounding residues. The hypericin molecules also set up contacts within themselves, interacting through hydrogen bonds established between their hydroxyl groups (Fig. 3 F). However, the higher affinity showed by phthalocyanine for the HR1 regions suggests that hydrophobic interactions can drive the formation of extremely favourable associations inside the pocket, hypothesis supported by the presence of many aromatic moieties in all the compounds showing higher affinity for this internal cavity (Fig. 2).
It is difficult to hypothesize what the location of these compounds might be in an intermediate or postfusion form of the spike, since complete postfusion structures are still not available. From a recently published incomplete structure of the spike postfusion core (PDB ID: 6lxt) (Xia et al., 2020) we found that only three of the key cavity residues, involved in interactions with the compounds, are present in the spike 6-helix bundle structure (residues 912–914). Nevertheless, since our aim was to prevent virus entry in an earlier phase, directly blocking the prefusion conformation, potential interactions of the drugs with structures in conformational progress were not taken into further consideration since they would not add details to improve our model.
3.3. MD simulations and MM/GBSA interaction energies evaluation
To further assess the affinity of the top two drugs for the spike pocket we performed 30 ns MD simulations of both multiple docking complexes to accurately evaluate, through the MM/GBSA method (Genheden and Ryde, 2015), the interaction energies between the docked compounds and the receptor. MM/GBSA analysis confirmed the high affinity of both drugs for the binding pocket (Table 2 ). In particular, MM/GBSA results highlighted that Van der Waals, electrostatic and non-polar solvation energy terms favourably contribute to the binding energy. The Van der Waals term, accounting the strength of nonpolar interactions derived from the binding of the drugs to the pocket, gives the highest contribution to the binding energy, showing extremely negative values. On the other hand, highly positive polar solvation energy terms have been obtained in all the calculations. This term, representing the energy associated with dissolving these drugs in the solvent, depict their preference for a hydrophobic environment.
Table 2.
Results of the MM/GBSA analyses performed for each of the three phthalocyanine and hypericin molecules within the binding pocket of the spike glycoprotein.
| Compound | VdW (kcal/mol) | Electrostatic (kcal/mol) | Nonpolar solvation (kcal/mol) | Polar solvation (kcal/mol) | Binding energy (kcal/mol) |
|---|---|---|---|---|---|
| phthalocyanine #1 | −84.8 | −5.6 | −6.8 | +30.6 | −66.6 |
| phthalocyanine #2 | −76.5 | −0.7 | −6.7 | +24.7 | −59.1 |
| phthalocyanine #3 | −54.3 | +3.0 | −5.0 | +20.7 | −35.6 |
| hypericin #1 | −55.3 | −18.4 | −4.7 | +34.7 | −43.6 |
| hypericin #2 | −53.5 | −19.4 | −5.2 | +33.8 | −44.1 |
| hypericin #3 | −37.4 | −14.4 | −4.3 | +28.8 | −27.3 |
Highly favourable Van der Waals energies have been obtained for the phthalocyanine drugs, while electrostatic interactions with surrounding residues play only a marginal role indicating that the massive hydrophobic contacts established by these molecules within the pocket are crucial for the stability of the complex. On the other hand, electrostatic interactions are important for the stability of the hypericin drugs, although the hydrophobic interactions still play a significant role for their binding affinity.
Hydrogen bond analysis, performed for both the simulations, further supports these results, showing that the presence of the three hypericin drugs leads to an increase in the hydrogen bonds network established within the pocket. In fact, these molecules can stably set up from 1 to 4 hydrogen bonds (Fig. S1, lower panel), mainly with residues P1090, R1091 and E1092 of the monomer A, N907, E1092 and Q1113 of the monomer B and P1090 of the monomer C, as well as with themselves. Conversely, polar interactions play only a minor role in phthalocyanine drugs binding to the pocket, since none of the three molecules established valuable hydrogen bonds during the simulation time (Fig. S1, upper panel).
Per-residue energy decomposition analysis (Genheden and Ryde, 2015) was also carried out for each drug to highlight the independent contribution of specific pocket residues to the binding, and to dynamically describe the compounds interactions within the pocket (Table S2). The analysis showed that the drugs can establish from 9 to 15 medium- to high-affinity contacts with residues belonging to the internal HR1 regions (residues 897–920) and with other surrounding residues inside the cavity. This evidence further supports the hypothesis that the drugs may act like anchors between surrounding internal regions. Moreover, the results also confirmed that the interactions established by the three phthalocyanines are mostly hydrophobic, while the three hypericin molecules can also set up valuable electrostatic interactions with most of the contacted residues, although the hydrophobic contribution is still much stronger.
The MD and MM/GBSA results mainly confirmed the interaction patterns described by the molecular docking simulations, indicating that highly stable binding poses have been obtained. These molecules improve the number of hydrophobic interactions inside the pocket, creating a non-polar network and the establishment of strong steric hindrance within the cavity that may likely hamper the subsequent conformational changes leading to the formation of the postfusion conformation of the spike. Interestingly, during the simulations it has been also observed that the first and second docked phthalocyanine or hypericin molecules maintain their starting binding poses interacting with the HR1 internal regions of the monomer A and B, while the third added phthalocyanine or hypericin compound moves towards the centre of the pocket, simultaneously contacting all the HR1 internal regions (Fig. 4 A–B). The arrangement of these compounds is expected to create a hard impairment to the motion of the HR1 regions, further contributing to the stabilization of the spike prefusion conformation. In conclusion, all these results sustain the hypothesis that the presence of these drugs inside the pocket may block the HR1 regions, preventing the huge spike conformational changes that permit the SARS-CoV-2 entry into the target cells.
Fig. 4.
Binding poses of the three phthalocyanine (A) and the three hypericin (B) molecules inside the spike glycoprotein pocket during the 30 ns MD simulations. Drugs are shown as spheres, coloured by atom type, with carbon atoms represented in grey. The spike glycoprotein is shown in blue cartoon representation with the HR1 regions highlighted in red. These images have been created using the UCSF Chimera 1.13.1 program (Pettersen et al., 2004).
3.4. Virtual screening ranking reweighting
The ranking of the compounds obtained through the VS procedure (Table S1) has been reweighted based on the literature retrieved for each compound (Table 3 , and Fig. S2 and S3). In particular, we conceived a scoring method which allowed us to evaluate a reformulated ranking based on: a) the obtained binding energies, b) known side effects and physiological effects of compounds and c) previously identified antiviral properties.
Table 3.
Final virtual screening ranking, obtained after applying the reweighting scoring function. Binding energies are calculated as an average from 3 repeated molecular docking simulations.
| Name | S(SE) | S(PE) | S(Trials) | S(Antiviral) | S(Binding) | S(VS) | S(final) |
|---|---|---|---|---|---|---|---|
| phthalocyanine | 4 | 0 | 5.3 | 35 | 16.3 | 2 | 62.6 |
| hypericin | 2 | 3 | 8.2 | 25 | 15.1 | 2 | 55.3 |
| TMC-647055 | 5 | 5 | 10.0 | 15 | 12.5 | 2 | 49.5 |
| quarfloxin | 5 | 5 | 5.6 | 5 | 12.6 | 2 | 35.2 |
| tepotinib | 5 | 0 | 7.1 | 0 | 12.0 | 0 | 24.1 |
| laniquidar | 5 | 0 | 5.2 | 0 | 12.8 | 0 | 23.0 |
| tadalafil | 2 | −3 | 10.0 | 0 | 12.4 | 0 | 21.4 |
| ergotamine | −1 | 0 | 5.9 | 0 | 13.2 | 2 | 20.1 |
| JNJ-10311795 | 1 | 5 | 0.0 | 0 | 13.1 | 0 | 19.1 |
| TZ2PA6 | 0 | 0 | 0.0 | 0 | 12.7 | 0 | 12.7 |
Briefly, side effects have been classified using a score ranging between -5 and +5 (named S(SE)). In particular, compounds interfering with other drugs have been penalized, compounds with mild side effects have been assigned a positive score and compounds with unknown effects received a score of 0. Physiological effects of the compounds also received a score ranging between -5 and 5 (named S(PE)), according to which compounds with unfavourable physiological activities were negatively weighted, compounds with predicted or evaluated antiviral or anti-inflammatory activity received a value between +1 and +5 and compounds with unknown or neutral physiological activities obtained a value of 0.
Drugs previously evaluated in clinical trials have been positively weighted, computing the clinical trial score (named S(Trials)) as follows:
| S(Trials) = 5+(1 x nAT)+(0.1 x nT)+(0.1 x nPhaseII)+(0.2 x nPhaseIII)+(0.3 x nPhaseIV) |
Where nAT indicates the number of clinical trials where the compound has been evaluated for its antiviral action, nT the number of trials with other clinical targets and nPhaseII, nPhaseIII, nPhaseIV, the number of trial in each experimental phase, as reported on the U.S. National Library of Medicine (NLM) Clinical Trials database (https://clinicaltrials.gov). A maximum value of 0.5, 1.0 and 1.5 was allowed for the last three terms of the equation to balance the ranking, and a maximum value of 10 for the overall S(Trials) score. Compounds with antiviral properties, even if directed towards viruses and/or targets different from the one chosen in our research, have been highly favoured in the final score. This property can add an extra value to the drugs, assuming that they could affect other SARS-CoV-2 targets in addition to the spike cavity. Furthermore, in particular in the case of phthalocyanine, the antiviral activity was experimentally observed but the specific viral targets were not completely understood (Vzorov et al., 2003), so the drug interaction within the inner cavity could provide a reason for this still undefined antiviral activity. For the S(Antiviral) calculation, compounds with known antiviral activity have been assigned a score of +10, while compounds for which this activity was only hypothesized, but not experimentally assessed, were assigned a score of +5. If the compound showed activity on enveloped viruses, a +5 was added to the score. Finally, according to confirmed data, a score between +10 and +20 was given to those compounds for which the reported inhibitory mechanism involved the binding of spike glycoproteins and/or the inhibition of virus fusion to host cells. For each compound, the average binding energy (named S(Binding)) has been calculated from three repeated molecular docking simulations and summed to the obtained score. Finally, a +2 wt was added to the best 5 compounds obtained from the VS procedure (named S(VS)), as reported in Table S1. Concluding, the final score for each compound was computed using the following equation:
| S(final)=S(SE)+S(PE)+S(Trials)+S(Antiviral)+S(Binding)+S(VS) |
The reweighted ranking reported in Table 3 shows that four top ranking compounds (i.e. phthalocyanine, hypericin, TMC-647055 and quarfloxin), evaluated combining of all the analyzed data, represent the most promising drugs and are suggested as a possible treatment against SARS-CoV-2 infection.
3.5. Known or hypothesized antiviral properties of the suggested compounds
To highlight the clinical potential of the four selected compounds, we provide a brief description for each drug, strictly focused on known or hypothesized antiviral properties. The listed effects, although not directed towards SARS-CoV-2 and not representing an evidence of interaction with this virus, may contribute to the general antiviral properties of the drug cocktail.
- Phthalocyanines are dye molecules commonly used for optical, electrical and catalytic applications (Sorokin, 2013; Wright, 2001). Different phthalocyanine derivatives have also been evaluated for their efficacy as virucidal agents and as fusion inhibitors (François et al., 2009; Vzorov et al., 2003, 2007). Given their photochemical properties, pththalocyanines also showed a phototoxic virucidal action on different enveloped viruses (Korneev et al., 2019; Nikolaeva-Glomb et al., 2017; Smetana et al., 1994). Concerning the safety of this molecule, no cell toxicity has been reported and an in vivo study on mice showed a good toxicity profile and no tissue damage (Styczynski et al., 2015).
- Hypericin is a natural naphthodiantrone compound mainly derived from plants of the genus Hypericum, particularly Hypericum perforatum (St. John’s Wort), and widely known for its antidepressant activity (Jendželovská et al., 2016; Karioti and Bilia, 2010). Hypericin has also shown specific antiviral activity on a wide variety of enveloped viruses, mainly related on the viral assembly and release inhibition (Kubin et al., 2005). Hypericin was also reported to act as a fusion inhibitor for different viruses (Kubin et al., 2005; Lenard et al., 1993; Stevenson and Lenard, 1993). Hypericin and H. perforatum extracts have also been shown to inhibit the cellular apoptosis induced by the avian coronavirus IBV, and to attenuate the pathological injuries caused by this virus in the trachea and kidney of chickens (Chen et al., 2019a, b). Side effects reported for St. John’s Wort treatment include skin rash, dizziness, headache, tiredness and difficult sleep.
- TMC-647055 is a macrocyclic indole, non-nucleoside inhibitor of the hepatitis C virus (HCV) NS5B RNA-dependent RNA Polymerase (Cummings et al., 2014; Devogelaere et al., 2012; Vendeville et al., 2012). Clinical trials have highlighted its promising antiviral properties against HCV, as well as its good safety and tolerability profiles, with limited toxicity and off-target activities. In combination with the TMC435 protease inhibitor (also known as simeprevir) and ritonavir showed an increased antiviral activity against HCV and was well tolerated by patients (Bourgeois et al., 2017; Vijgen et al., 2017).
- Quarfloxin (also known as CX-3543 or itarnafloxin), a fluoroquinolone derivative, is a G-quadruplex-binding compound, which has been clinically evaluated for its antineoplastic activity (Balasubramanian et al., 2011; Drygin et al., 2009). This compound was well tolerated by patients during clinical trials and showed a good toxicity profile, but after reaching Phase II it was discarded for poor bioavailability at the action site (Balasubramanian et al., 2011). Other G-quadruplex binding compounds were shown to exert antiviral activity against different enveloped viruses and to interact with G-quadruplex regions of viral genomes (Lavezzo et al., 2018; Ruggiero and Richter, 2018). For this reason, we hypothesize that quarfloxin could possess a potential antiviral activity, despite to our knowledge it has not yet been evaluated.
3.6. Spike glycoproteins sequence alignments
The inhibition mechanism hypothesized in this research strictly depends on the interactions established between the compounds and the HR1 internal residues of the pocket. This mechanism may be consequently extended to other members of the betacoronavirus family depending on the sequence identity found in this region. With this purpose, the sequence of the spike glycoprotein structure used in this study has been first checked against the high number of SARS-CoV-2 sequences deposited until April 1, 2020 in the GenBank Protein database (Clark et al., 2016). We retrieved all complete SARS-CoV-2 spike glycoprotein protein sequences from the NCBI Virus portal (Hatcher et al., 2017), and performed a similarity search using the blast-p suite of BLAST (Altschul et al., 1990), selecting only the region spanning from the fusion peptide to the HR1 region (residues from 811 to 984 in the cryo-EM structure). The 174-residues long query sequence showed 100 % identity towards 345 of the 360 evaluated spike-glycoprotein target sequences, indicating that this region is highly conserved between all the SARS-CoV-2 isolates sequenced to date. Among 15 entries with an identity lower than 100 %, two sequences showed only one residue mismatch, located outside of the internal cavity, while the others contained undetermined residues that do not match to the query, explaining the lower identity percentage obtained. We further evaluated whether this region was also conserved among other betacoronaviruses (NCBI:txid694002). The submitted query sequence showed a 100 % identity with the spike glycoprotein sequence of the bat coronavirus RaTG13 (NCBI:txid2709072; GenBank accession: QHR63300.2). Identity values ranging from about 88 to 95 % were observed for different SARS-CoV isolates and other animal SARS-like betacoronaviruses, for a total of 148 different entries. In particular, the internal HR1 region, the specific target of our compounds, is completely conserved among all these 148 hit sequences, except for one residue mismatch observed within the sequence of the palm civet SARS coronavirus A013 (NCBI:txid305402; GenBank accession: AAV97985.1). On the other hand, the spike glycoprotein sequences deposited for the middle east respiratory syndrome coronavirus (MERS-CoV), another concerning human pathogen, only showed about 52–57% sequence identity against the fusion peptide-HR1 SARS-CoV-2 region, and an identity between about 58 and 67 % for the internal HR1 region alone.
These results show that our selected compounds, targeting the highly conserved internal regions of the spike, could represent a broad-spectrum treatment active against all SARS-CoV-2 currently isolated variants, overcoming the concerning mutation potential of this virus. Furthermore, the high sequence and structural identities between the SARS-CoV and the SARS-CoV-2 spike glycoproteins suggest that these compounds may represent an efficient therapeutic strategy also towards the SARS-CoV virus. Finally, the low sequence identity evaluated between the SARS-CoV-2 internal HR1 region and that of other members of the betacoronavirus family do not completely exclude the possibility that these drugs may also effectively bind within these non-identical pockets, since the different pattern of interactions that would be established could perhaps enhance the binding of the compounds, instead of preventing it.
4. Conclusions
The results obtained in this research support the hypothesis that the presence of specific compounds inside the inner cavity of the spike glycoprotein in prefusion conformation (Fig. 1) could block the HR1 regions, preventing the conformational changes that allow SARS-CoV-2 entry into target cells. In fact, the HR1 regions are actively involved in the compounds recognition, as confirmed by the MM/GBSA per-residue decomposition analysis (Table S2). In addition, our hypothesized inhibitory mechanism may explain the unknown mechanisms by which phthalocyanine derivatives have been experimentally shown to induce viral fusion inhibition with no impairment in spike glycoproteins binding to cell receptors (Vzorov et al., 2003).
We suggest that an optimal therapeutic strategy could involve the simultaneous administration of a cocktail of inhibitors that could boost up each individual antiviral property. Based on the reweighted virtual screening results (Table 3), obtained though the evaluation of the previous literature, we propose the concurrent administration of four compounds: phthalocyanine, hypericin, TMC-647055 and quarfloxin, to be used as a possible antiviral treatment against SARS-CoV-2. Their already observed antiviral properties, although mostly assessed on different targets and viruses, add an important value to these drugs, since they could interfere with SARS-CoV-2 infection also with other mechanisms independent from the hypothesized inhibition of spike glycoprotein fusion to the host cell.
To evaluate the feasibility of the simultaneous localization of different compounds inside the pocket we also performed sequential molecular docking simulations of these four selected drugs (Fig. S4). Results showed that the compounds could fit together inside the pocket still maintaining a high affinity for the HR1 regions and the overall binding pocket, where evaluated interaction energies are: phthalocyanine -16.7, hypericin -13.7, TMC-647055 -12.7 and quarfloxin -11.6 kcal/mol.
Furthermore, the results obtained carrying out the sequence alignments strongly indicate that the hypothesized inhibition mechanism described in this work may be extended to all the identified SARS-CoV-2 isolates, to the closely related SARS-CoV, and potentially even to other known members of the betacoronavirus family.
Concluding, the hypotheses arising from this simulation research can represent a valid contribution to the international community for the modulation of the SARS-CoV-2 infection, and could also be useful against other enveloped viruses sharing the same infection mechanism.
Author contributions
M.F., F.I. and A.R. conceived, designed and wrote the paper; A.R. and F.I. performed the calculations and analyzed the data.
CRediT authorship contribution statement
Alice Romeo: Conceptualization, Writing - original draft, Formal analysis, Investigation. Federico Iacovelli: Conceptualization, Writing - original draft, Formal analysis, Investigation. Mattia Falconi: Project administration, Conceptualization, Methodology, Writing - original draft.
Declaration of Competing Interest
The authors declare they have no potential conflicts of interest to disclose.
Acknowledgments
The computing resources and the related technical support were provided by CRESCO/ENEAGRID High Performance Computing infrastructure (Ponti et al., 2014). CRESCO/ENEAGRID High Performance Computing infrastructure is funded by ENEA, the Italian National Agency for New Technologies, Energy and Sustainable Economic Development and by Italian and European research programmes, see http://www.cresco.enea.it/english for information.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.virusres.2020.198068.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindah E. Gromacs: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1-2:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Aoki K.M., Yoneya M., Yokoyama H. Constant pressure Md simulation method. Mol. Cryst. Liq. Cryst. 2004;413(1):109–116. doi: 10.1080/15421400490437259. [DOI] [Google Scholar]
- Balasubramanian S., Hurley L.H., Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat. Rev. Drug Discov. 2011;10(4):261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battles M.B., McLellan J.S. Respiratory syncytial virus entry and how to block it. Nat. Rev. Microbiol. 2019;17:233–245. doi: 10.1038/s41579-019-0149-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battles M.B., Langedijk J.P., Furmanova-Hollenstein P., Chaiwatpongsakorn S., Costello H.M., Kwanten L., Vranckx L., Vink P., Jaensch S., Jonckers T.H., Koul A., Arnoult E., Peeples M.E., Roymans D., McLellan J.S. Molecular mechanism of respiratory syncytial virus fusion inhibitors. Nat. Chem. Biol. 2016;12(2):87–93. doi: 10.1038/nchembio.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B.J., van der Zee R., de Haan C.A., Rottier P.J. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77(16):8801–8811. doi: 10.1128/jvi.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois S., Van Vlierberghe H., Moreno C., Orlent H., Nevens F., Arastéh K., Horsmans Y., Schattenberg J.M., Buggisch P., Francque S., Vijgen L., Kakuda T.N., Hoeben E., Luo D., Vandebosch A., Jacquemyn B., Van Remoortere P., Verloes R. Efficacy, safety and pharmacokinetics of simeprevir and TMC647055/ritonavir with or without ribavirin and JNJ-56914845 in HCV genotype 1 infection. BMC Gastroenterol. 2017;17(1):26. doi: 10.1186/s12876-017-0580-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case D.A., Ben-Shalom I.Y., Brozell S.R., Cerutti D.S., Cheatham III T.E., Cruzeiro V.W.D., Darden T.A., Duke R.E., Ghoreishi D., Gilson M.K., Gohlke H., Goetz A.W., Greene D., Harris R., Homeyer N., Izadi S., Kovalenko A., Kurtzman T., Lee T.S., LeGrand S., Li P., Lin C., Liu J., Luchko T., Luo R., Mermelstein D.J., Merz K.M., Miao Y., Monard G., Nguyen C., Nguyen H., Omelyan I., Onufriev A., Pan F., Qi R., Roe D.R., Roitberg A., Sagui C., Schott-Verdugo S., Shen J., Simmerling C.L., Smith J., Salomon-Ferrer R., Swails J., Walker R.C., Wang J., Wei H., Wolf R.M., Wu X., Xiao L., York D.M., Kollman P.A. University of California; San Francisco: 2018. AMBER 2018. [Google Scholar]
- Chen H., Muhammad I., Zhang Y., Ren Y., Zhang R., Huang X., Diao L., Liu H., Li X., Sun X., Abbas G., Li G. Antiviral activity against infectious bronchitis virus and bioactive components of Hypericum perforatum l. Front. Pharmacol. 2019;10:1272. doi: 10.3389/fphar.2019.01272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Feng R., Muhammad I., Abbas G., Zhang Y., Ren Y., Huang X., Zhang R., Diao L., Wang X., Li G. Protective effects of hypericin against infectious bronchitis virus induced apoptosis and reactive oxygen species in chicken embryo kidney cells. Poult. Sci. 2019;98(12):6367–6377. doi: 10.3382/ps/pez465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W. GenBank. Nucleic Acids Res. 2016;44(D1):D67–D72. doi: 10.1093/nar/gkv1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings M.D., Lin T.I., Hu L., Tahri A., McGowan D., Amssoms K., Last S., Devogelaere B., Rouan M.C., Vijgen L., Berke J.M., Dehertogh P., Fransen E., Cleiren E., van der Helm L., Fanning G., Nyanguile O., Simmen K., Van Remoortere P., Raboisson P., Vendeville S. Discovery and early development of TMC647055, a non-nucleoside inhibitor of the hepatitis C virus NS5B polymerase. J. Med. Chem. 2014;57(5):1880–1892. doi: 10.1021/jm401396p. [DOI] [PubMed] [Google Scholar]
- Darden T., York D., Pedersen L. Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98(12):10089. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- de Wit E., Feldmann F., Cronin J., Jordan R., Okumura A., Thomas T., Scott D., Cihlar T., Feldmann H. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc. Natl. Acad. Sci. U.S.A. 2020;117(12):6771–6776. doi: 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVincenzo J.P., Whitley R.J., Mackman R.L., Scaglioni-Weinlich C., Harrison L., Farrell E., McBride S., Lambkin-Williams R., Jordan R., Xin Y., Ramanathan S., O’Riordan T., Lewis S.A., Li X., Toback S.L., Lin S.L., Chien J.W. Oral GS-5806 activity in a respiratory syncytial virus challenge study. N. Engl. J. Med. 2014;371(8):711–722. doi: 10.1056/NEJMoa1401184. [DOI] [PubMed] [Google Scholar]
- Devogelaere B., Berke J.M., Vijgen L., Dehertogh P., Fransen E., Cleiren E., van der Helm L., Nyanguile O., Tahri A., Amssoms K., Lenz O., Cummings M.D., Clayton R.F., Vendeville S., Raboisson P., Simmen K.A., Fanning G.C., Lin T.I. TMC647055, a potent nonnucleoside hepatitis C virus NS5B polymerase inhibitor with cross-genotypic coverage. Antimicrob. Agents Chemother. 2012;56(9):4676–4684. doi: 10.1128/AAC.00245-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drygin D., Siddiqui-Jain A., O’Brien S., Schwaebe M., Lin A., Bliesath J., Ho C.B., Proffitt C., Trent K., Whitten J.P., Lim J.K., Von Hoff D., Anderes K., Rice W.G. Anticancer activity of CX-3543: a direct inhibitor of rRNA biogenesis. Cancer Res. 2009;69(19):7653–7661. doi: 10.1158/0008-5472.CAN-09-1304. [DOI] [PubMed] [Google Scholar]
- François K.O., Pannecouque C., Auwerx J., Lozano V., Pérez-Pérez M.J., Schols D., Balzarini J. The phthalocyanine prototype derivative Alcian Blue is the first synthetic agent with selective anti-human immunodeficiency virus activity due to its gp120 glycan-binding potential. Antimicrob. Agents Chemother. 2009;53(11):4852–4859. doi: 10.1128/AAC.00811-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genheden S., Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015;10(5):449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goga N., Rzepiela A.J., de Vries A.H., Marrink S.J., Berendsen H.J.C. Efficient algorithms for Langevin and DPD dynamics. J. Chem. Theory Comput. 2012;8(10):3637–3649. doi: 10.1021/ct3000876. [DOI] [PubMed] [Google Scholar]
- Hatcher E.L., Zhdanov S.A., Bao Y., Blinkova O., Nawrocki E.P., Ostapchuck Y., Schäffer A.A., Brister J.R. Virus Variation Resource - improved response to emergent viral outbreaks. Nucleic Acids Res. 2017;45(D1):D482–D490. doi: 10.1093/nar/gkw1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Jendželovská Z., Jendželovský R., Kuchárová B., Fedoročko P. Hypericin in the light and in the dark: two sides of the same coin. Front. Plant Sci. 2016;7:560. doi: 10.3389/fpls.2016.00560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79(2):926. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- Karioti A., Bilia A.R. Hypericins as potential leads for new therapeutics. Int. J. Mol. Sci. 2010;11(2):562–594. doi: 10.3390/ijms11020562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneev D., Kurskaya O., Sharshov K., Eastwood J., Strakhovskaya M. Ultrastructural aspects of photodynamic inactivation of highly pathogenic avian H5N8 influenza virus. Viruses. 2019;11(10):955. doi: 10.3390/v11100955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubin A., Wierrani F., Burner U., Alth G., Grünberger W. Hypericin - the facts about a controversial agent. Curr. Pharm. Des. 2005;11(2):233–253. doi: 10.2174/1381612053382287. [DOI] [PubMed] [Google Scholar]
- Kupferschmidt K., Cohen J. Race to find COVID-19 treatments accelerates. Science. 2020;367(6485):1412–1413. doi: 10.1126/science.367.6485.1412. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., Swindells M.B. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011;51(10):2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- Lavezzo E., Berselli M., Frasson I., Perrone R., Palù G., Brazzale A.R., Richter S.N., Toppo S. G-quadruplex forming sequences in the genome of all known human viruses: a comprehensive guide. PLoS Comput. Biol. 2018;14(12) doi: 10.1371/journal.pcbi.1006675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenard J., Rabson A., Vanderoef R. Photodynamic inactivation of infectivity of human immunodeficiency virus and other enveloped viruses using hypericin and rose bengal: inhibition of fusion and syncytia formation. Proc. Natl. Acad. Sci. U.S.A. 1993;90(1):158–162. doi: 10.1073/pnas.90.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3(1):237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes B.R.P., da Costa M.F., Genova Ribeiro A., da Silva T.F., Lima C.S., Caruso I.P., de Araujo G.C., Kubo L.H., Iacovelli F., Falconi M., Desideri A., de Oliveira J., Regasini L.O., de Souza F.P., Toledo K.A. Quercetin pentaacetate inhibits in vitro human respiratory syncytial virus adhesion. Virus Res. 2020;276 doi: 10.1016/j.virusres.2019.197805. [DOI] [PubMed] [Google Scholar]
- Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaeva-Glomb L., Mukova L., Nikolova N., Kussovski V., Doumanova L., Mantareva V., Angelov I., Wöhrle D., Galabov A.S. Photodynamic effect of some phthalocyanines on enveloped and naked viruses. Acta Virol. 2017;61(3):341–346. doi: 10.4149/av_2017_313. [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera -- a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Ponti G., Palombi F., Abate D., Ambrosino F., Aprea G., Bastianelli T., Beone F., Bertini R., Bracco G., Caporicci M., Calosso B., Chinnici M., Colavincenzo A., Cucurullo A., Dangelo P., De Rosa M., De Michele P., Funel A., Furini G., Giammattei D., Giusepponi S., Guadagni R., Guarnieri G., Italiano A., Magagnino S., Mariano A., Mencuccini G., Mercuri C., Migliori S., Ornelli P., Pecoraro S., Perozziello A., Pierattini S., Podda S., Poggi F., Quintiliani A., Rocchi A., Scio C., Simoni F., Vita A. The role of medium size facilities in the HPC ecosystem: the case of the new CRESCO4 cluster integrated in the ENEAGRID infrastructure. Proceedings of the 2014 International Conference on High Performance Computing and Simulation (HPCS) 2014:1030–1033. doi: 10.1109/HPCSim.2014.6903807. art. no. 6903807. [DOI] [Google Scholar]
- Ruggiero E., Richter S.N. G-quadruplexes and G-quadruplex ligands: targets and tools in antiviral therapy. Nucleic Acids Res. 2018;46(7):3270–3283. doi: 10.1093/nar/gky187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckaert J.P., Ciccotti G., Berendsen H.J.C. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of N-Alkanes. J. Comput. Phys. 1977;23(3):327–341. doi: 10.1016/0021-9991(77)90098-5. [DOI] [Google Scholar]
- Samuel D., Xing W., Niedziela-Majka A., Wong J.S., Hung M., Brendza K.M., Perron M., Jordan R., Sperandio D., Liu X., Mackman R., Sakowicz R. GS-5806 inhibits pre- to postfusion conformational changes of the respiratory syncytial virus fusion protein. Antimicrob. Agents Chemother. 2015;59(11):7109–7112. doi: 10.1128/AAC.00761-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanner M.F. Python: a programming language for software integration and development. J. Mol. Graph. Model. 1999;17(1):57–61. [PubMed] [Google Scholar]
- Smetana Z., Mendelson E., Manor J., van Lier J.E., Ben-Hur E., Salzberg S., Malik Z. Photodynamic inactivation of herpes viruses with phthalocyanine derivatives. J. Photochem. Photobiol. B. 1994;22(1):37–43. doi: 10.1016/1011-1344(93)06949-4. [DOI] [PubMed] [Google Scholar]
- Sorokin A.B. Phthalocyanine metal complexes in catalysis. Chem. Rev. 2013;113(10):8152–8191. doi: 10.1021/cr4000072. [DOI] [PubMed] [Google Scholar]
- Stevens M., Rusch S., DeVincenzo J., Kim Y.I., Harrison L., Meals E.A., Boyers A., Fok-Seang J., Huntjens D., Lounis N., Mariёn K., Remmerie B., Roymans D., Koul A., Verloes R. Antiviral activity of oral JNJ-53718678 in healthy adult volunteers challenged with respiratory syncytial virus: a placebo-controlled study. J. Infect. Dis. 2018;218(5):748–756. doi: 10.1093/infdis/jiy227. [DOI] [PubMed] [Google Scholar]
- Stevenson N.R., Lenard J. Antiretroviral activities of hypericin and rose bengal: photodynamic effects on Friend leukemia virus infection of mice. Antiviral Res. 1993;21(2):119–127. doi: 10.1016/0166-3542(93)90048-n. [DOI] [PubMed] [Google Scholar]
- Styczynski A.R., Anwar K.N., Sultana H., Ghanem A., Lurain N., Chua A., Ghassemi M., Novak R.M. In vitro antiretroviral activity and in vivo toxicity of the potential topical microbicide copper phthalocyanine sulfate. Virol. J. 2015;12:132. doi: 10.1186/s12985-015-0358-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The PyMOL Molecular Graphics System Version 2.1.0 Schrödinger, LLC.
- Tian C., Kasavajhala K., Belfon K.A.A., Raguette L., Huang H., Migues A.N., Bickel J., Wang Y., Pincay J., Wu Q., Simmerling C. ff19SB: amino-acid-Specific protein backbone parameters trained against quantum mechanics energy surfaces in solution. J. Chem. Theory Comput. 2020;16(1):528–552. doi: 10.1021/acs.jctc.9b00591. [DOI] [PubMed] [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendeville S., Lin T.I., Hu L., Tahri A., McGowan D., Cummings M.D., Amssoms K., Canard M., Last S., Van den Steen I., Devogelaere B., Rouan M.C., Vijgen L., Berke J.M., Dehertogh P., Fransen E., Cleiren E., van der Helm L., Fanning G., Van Emelen K., Nyanguile O., Simmen K., Raboisson P. Finger loop inhibitors of the HCV NS5b polymerase. Part II. Optimization of tetracyclic indole-based macrocycle leading to the discovery of TMC647055. Bioorg. Med. Chem. Lett. 2012;22(13):4437–4443. doi: 10.1016/j.bmcl.2012.04.113. [DOI] [PubMed] [Google Scholar]
- Vijgen L., Thys K., Vandebosch A., Van Remoortere P., Verloes R., De Meyer S. Virology analysis in HCV genotype 1-infected patients treated with the combination of simeprevir and TMC647055/ritonavir, with and without ribavirin, and JNJ-56914845. Virol. J. 2017;14(1):101. doi: 10.1186/s12985-017-0760-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vzorov A.N., Marzilli L.G., Compans R.W., Dixon D.W. Prevention of HIV-1 infection by phthalocyanines. Antiviral Res. 2003;59(2):99–109. doi: 10.1016/s0166-3542(03)00035-4. [DOI] [PubMed] [Google Scholar]
- Vzorov A.N., Bozja J., Dixon D.W., Marzilli L.G., Compans R.W. Parameters of inhibition of HIV-1 infection by small anionic microbicides. Antiviral Res. 2007;73(1):60–68. doi: 10.1016/j.antiviral.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Walls A.C., Tortorici M.A., Snijder J., Xiong X., Bosch B.J., Rey F.A., Veesler D. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc. Natl. Acad. Sci. U.S.A. 2017;114(42):11157–11162. doi: 10.1073/pnas.1708727114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Wolf R.M., Caldwell J.W., Kollman P.A., Case D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004;25(9):1157–1174. doi: 10.1002/jcc.20035. Erratum in: 2005. J. Comput. Chem. 26 (1), 114. [DOI] [PubMed] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., Lepore R., Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z., Assempour N., Iynkkaran I., Liu Y., Maciejewski A., Gale N., Wilson A., Chin L., Cummings R., Le D., Pon A., Knox C., Wilson M. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright J.D. Phthalocyanines. In: Buschow K.H.J., Cahn R.W., Flemings M.C., Ilschner B., Kramer E.J., Mahajan S., Veyssière P., editors. Encyclopedia of Materials: Science and Technology (Second Edition) Elsevier; 2001. pp. 6987–6991. [DOI] [Google Scholar]
- Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S., Qin C., Sun F., Shi Z., Zhu Y., Jiang S., Lu L. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30(4):343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D., Lee S., Thakkar V.D., Luo M., Moore M.L., Plemper R.K. Cross-resistance mechanism of respiratory syncytial virus against structurally diverse entry inhibitors. Proc. Natl. Acad. Sci. U.S.A. 2014;111(33):E3441–E3449. doi: 10.1073/pnas.1405198111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.