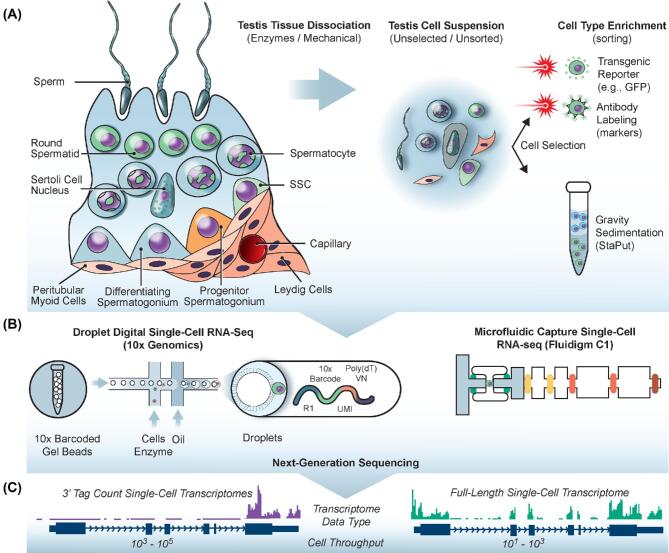
Figure 1.
Application of single cell RNA-seq to the study of spermatogenesis. (A) The complex testicular tissue is dissociated enzymatically and with mechanical force into individual cells. Many scRNA-seq studies explored the transcriptomes of these cell unselected/unsorted cell suspensions, while others used various strategies reliant on transgenic reporters, antibody labeling or cell density to enrich for particular cell types. (B) Unsorted or selected cell suspensions were then used for single-cell RNA-seq. Two popular platforms are the 10x Genomics Chromium (left) and Fluigidm C1 (right). In the 10x Genomics droplet-digital RNAseq approach, individual cells are capsulated in aqueous droplets together with microbeads that deliver barcoded primers for reverse transcription. The Fluidigm C1 physically captures individual cells on microfluidic chips and automatically generates full-length cDNA. (C) After next-generation sequencing of single-cell RNA-seq libraries, the 10x Genomics and Fluidigm C1 produce different data types depicted by the “tracks” of colored sequence located above the genome annotation. 10x Genomics libraries utilize 3′ end-counting chemistry, which maximizes cell throughput and better controls for PCR duplication bias through the use of unique molecular identifiers (UMIs). Fluidigm C1 libraries are full-length transcriptomes, meaning that mRNA variants (e.g., spliceoforms, alternative TSS usage) can be recognized, but with the drawback of reduced cell throughput and greater expense. This figure is available in color at Biology of Reproduction online.