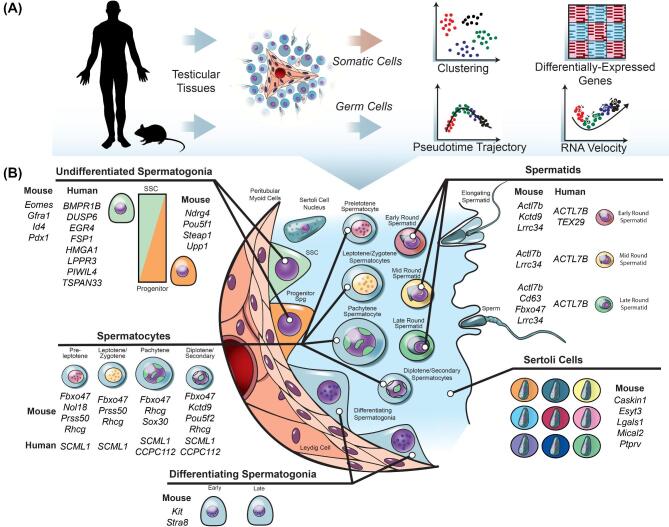
Figure 3.
Analyses and outcomes of scRNA-seq data from mouse and human spermatogenic cells. (A) Cells from human and mouse testicular tissue have been used for separate analyses of testicular germ and somatic cells at the single-cell level (Tables 1 & 2). In general, the analysis pipelines of various studies involve unbiased cell clustering, detection of differentially expressed genes (DEGs) between clusters, and pseudotime trajectory analysis to infer developmentally-regulated gene expression changes. (B) In summary, from the various scRNA-seq studies of spermatogenic cells, at least 11 distinct types of spermatogenic cells have been reported, distinguished by DEGs and 9 types of Sertoli cells. Noted are examples of validated genes corresponding to these various cell types. This figure is available in color at Biology of Reproduction online.