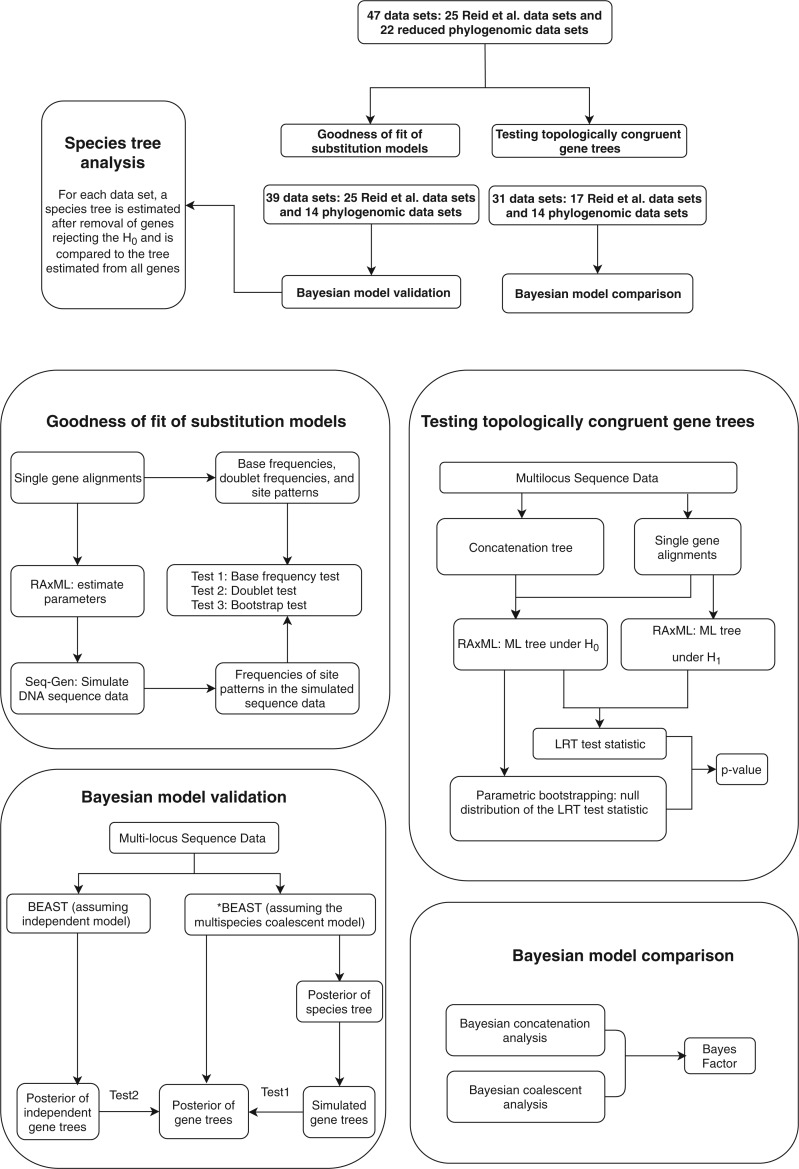
Figure 1.
Flowchart of phylogenetic analyses in this study. The alignments of 47 data sets were used to test the hypothesis of TC gene trees and goodness of fit of substitution models. Model validation and comparison were only performed for the loci that fit the GTRGAMMA model, reducing the number of input data for model validation to 39 (25 Reid et al. data sets and 14 phylogenomic data sets). Because model comparison as implemented in BEAST does not allow missing taxa in any locus, eight Reid et al. data sets were removed, further reducing the number of input data for model comparison to 31 (17 Reid et al. data sets and 14 phylogenomic data sets).