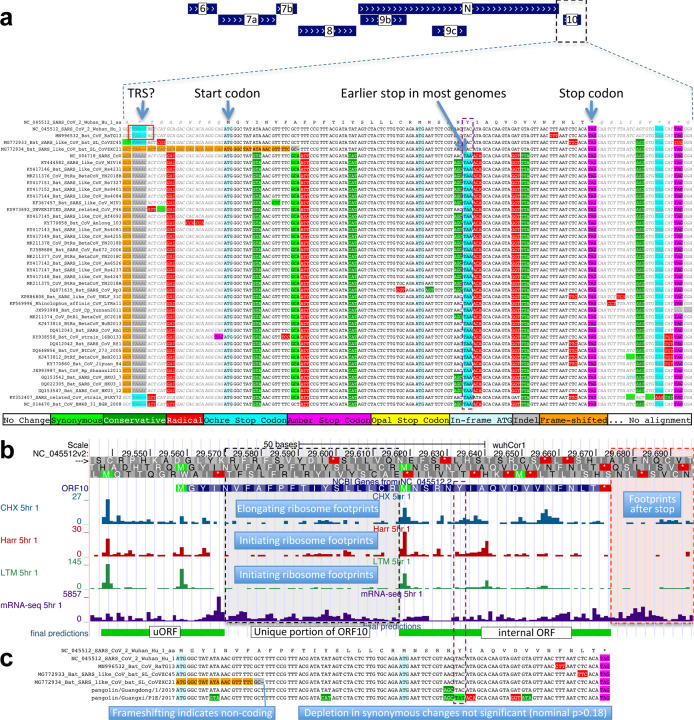
Extended Data Figure 3. ORF10 is not protein-coding.
a. Alignment of Sarbecovirus genomes at ORF10, including 30 additional flanking nucleotides on each side. Most substitutions are amino-acid-changing, either radical (red) or conservative (dark green), with only two synonymously-changing positions (light green), indicating this is not a protein-coding region. In addition, nearly all strains show an earlier stop codon (cyan), further reducing the length of this already-short ORF from 38 amino-acids to 25, and one of the four strains lacking the earlier stop includes a frame-shifting deletion. The putative partial transcription-regulatory sequence (TRS) present in SARS-CoV-2 and its closest relative (Bat CoV RaTG13) is not conserved in any other strains. The region surrounding ORF10 shows very high nucleotide-level conservation, which spans ORF10 and extends beyond its boundaries in both directions, indicating that this portion of the genome is functionally important even though it does not code for protein (indeed, this region is part of a pseudoknot RNA structure involved in RNA synthesis). b. Ribosome footprints previously used to suggest ORF10 translation10 in fact localize either in an upstream ORF (uORF, green) or in an internal ORF (green, “final predictions” track10), but not in the unique portion of ORF10 (dashed black box), indicating they are less likely to reflect functional translation of ORF10, and more likely to represent incidental translation initiation events. We note that the density of elongating footprints in the unique portion (black box) is no greater than the density after the stop codon (red box), consistent with incidental events. We also note that the internal ORF is only 18 codons long in 4 strains, and only 5 codons long in the other 40 Sarbecovirus strains, given the early stop codon (purple box) and unlikely to be functional. Footprint tracks show elongating ribosome footprints in cells treated with cycloheximide (blue, CHX), and footprints enriched for initiating ribosomes using harringtonine (Harr, red), and lactimidomycin (LTM, green). “mRNA-seq” track shows RNA-seq reads. c. CodAlignView25 of alignment previously used to argue that a high dN/dS ratio in ORF10 indicated positive selection for protein-coding-like rapid evolution8, based on only six closely-related strains (SARS-CoV-2, three bat viruses, two pangolin viruses). The authors noted a frameshifting deletion (orange/grey) in one of the bat viruses, which provides strong evidence against conserved protein-coding function, but they interpreted it (without evidence) as a potential sequencing error and excluded the strain from consideration. Even ignoring the frameshift-containing strain, the evidence used is insufficient to reach statistical significance: the alignment includes only 9 substitutions, of which 4 are radical, 4 are conservative, and 1 is synonymous. In a neutrally-evolving region with 9 substitutions, we would expect 2–3 synonymous changes, depending on the evolutionary model used, and a depletion to only 1 synonymous change is not statistically significant (nominal p-value>0.18 even in the most generous evolutionary model). This already-non-significant nominal p-value would move even further from significance with the necessary multiple-hypothesis corrections.