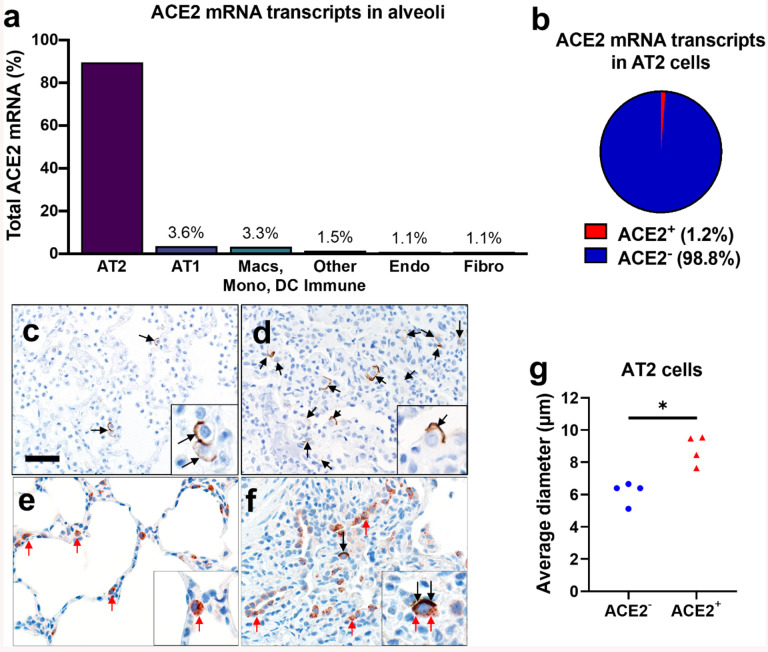
Figure 1. ACE2 expression in human lung.
a, b) Single-cell RNA sequencing reanalysis of ACE2 transcript abundance in alveoli from lung parenchyma samples (26). Summative observations from all donors. Airway cells (basal, mitotic, ciliated, club) are not shown. a) 89·5% of the cells with detectable ACE2 mRNA in the alveoli are alveolar type II cells. b) Only 1·2% of alveolar type II cells have ACE2 mRNA transcripts. c-f) Detection of ACE2 protein (brown color, black arrows, and insets) in representative sections of lower respiratory tract regions and tissue scoring (see Supplemental Table 2) (g). c, d) Alveolar regions had uncommon to regional polarized apical staining of solitary epithelial cells (c) that (when present) were more readily detected in collapsed regions of lung (d). e, f) SP-C (red arrows, inset) and ACE2 (black arrows, inset) dual immunohistochemistry on the same tissue sections. e) Non-collapsed regions had normal SP-C+ AT2 cells lacking ACE2. f) Focal section of peri-airway remodeling and collapse with several SP-C+ (red arrows) AT2 cells, but only a small subset of AT2 cells had prominent apical ACE2 protein (black arrows, inset). g) SP-C+/ACE2+ AT2 cells were often larger than SP-C+/ACE2− AT2 cells from same lung sections (see also d and e insets) indicative of AT2 hypertrophy, each data point represents the average value for each case from 5–10 cell measurements per group, P=0·0014, paired T-test. AT2: alveolar type II. AT1: alveolar type I. Macs: Macrophages. Mono: Monocytes. DC: dendritic cells. Other immune cells: B cells, mast cells, natural killer/T cells. Endo: Endothelial. Fibro: Fibroblasts/myofibroblasts. Bar = 35 μm.