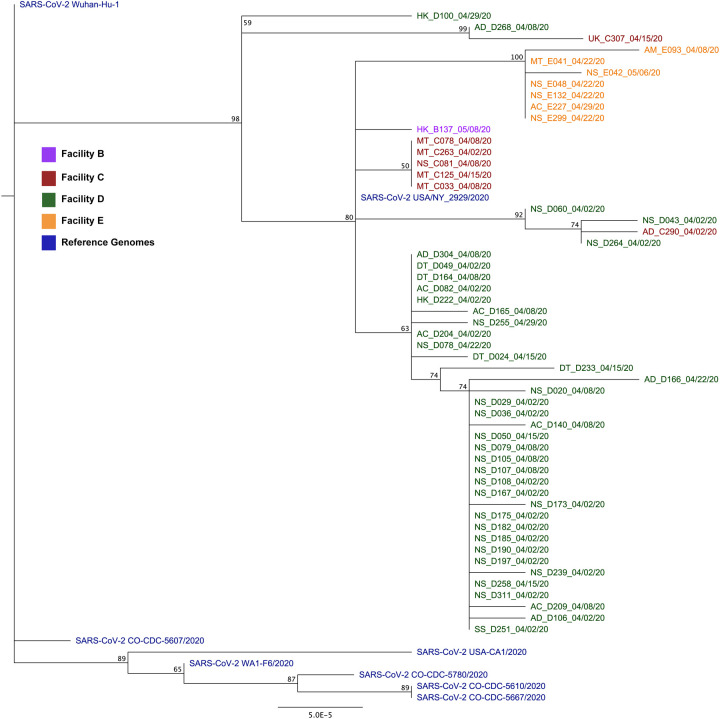
Figure 5. Phylogenetic analysis of SARS-CoV-2 genomes collected from Colorado LTCFs.
A) PhyML tree constructed using Tamura-Nei distance model including both transitions and transversions in Geneious Prime. Node numbers indicate bootstrap confidence based on 1000 replicates. Distance matrix was computed, and the tree was visualized in Geneious Prime. Letters at the beginning of taxon names represent job code (AC-activities, AD-administrative, AM-admissions, DT-dietary, MT-maintenance, NS-nursing, SS-social services, UK-unknown), and A-E letter indicate site of origin. Numbers after underscore indicate the date of sample collection. Reference sequences and four Colorado-derived sequences were obtained from NCBI. B) Map of the LTCFs’ relative geographic locations and distances from one another.