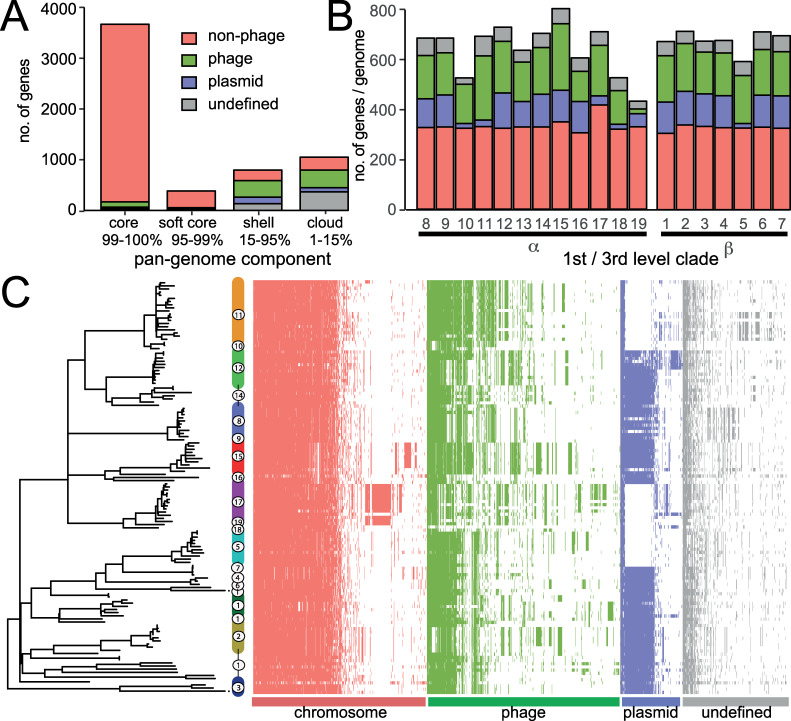
Fig 2. The pan genome of 131 S. Typhimurium isolates.
Gene families were identified based on sequence alignment with a cut off of 90% sequence identity and assigned to non-prophage chromosomal (red), prophage (green), plasmid (blue), or undefined (grey), based on their genome context in eleven annotated reference genomes from each third level clade. (A) Number of genome families in the core, softcore, shell and cloud components of the pangenome. (B) Number of genome families of each pan genome component in isolates from each S. Typhimurium third level clade. (C) Accessory genome (shell and cloud) in each isolate. Gene families present in more than 130 or less than 5 strains were excluded. Maximum Likelihood tree based on variation (SNPs) in the core genome with reference to S. Typhimurium SL1344. Third-level clades are indicated in colour coded in common with the phylogeny vertical bars.