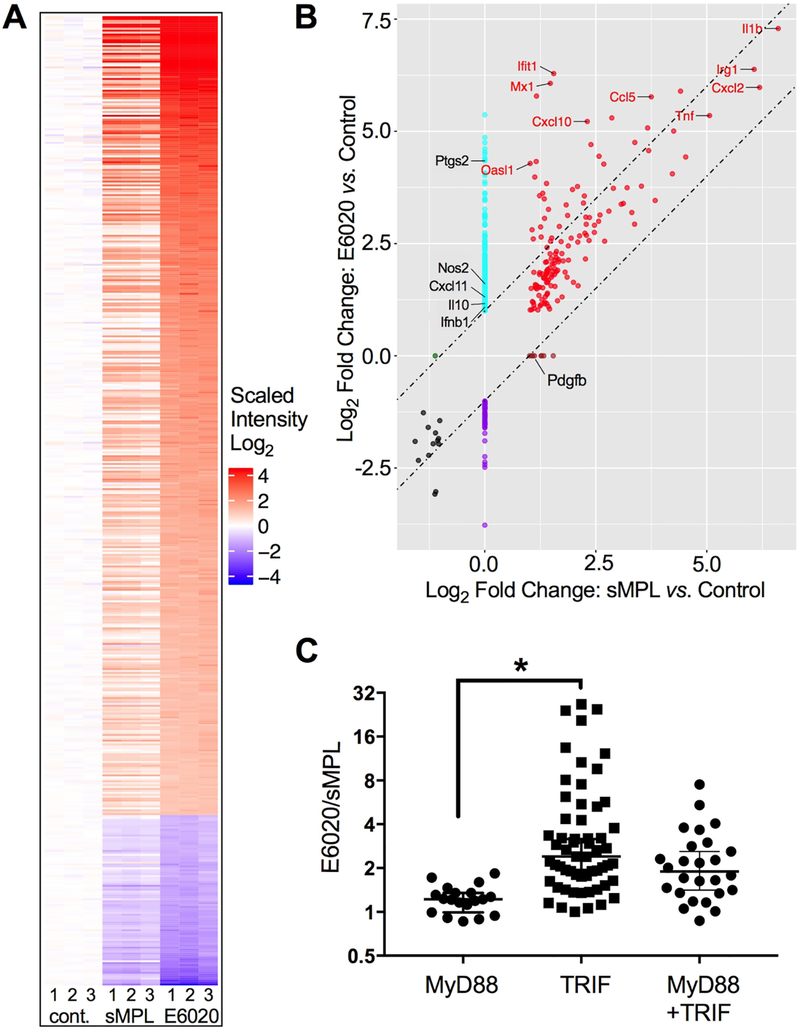
Figure 2. E6020 induces a strong Type I IFN signature.
Primary macrophages were either treated for 2 h with medium only, sMPL, or E6020 (100 ng/mL). RNA was subjected to microarray analysis. (A) Heatmap of differentially expressed (2-fold cut-off, FDR < 0.05) protein-coding and complex genes from 3 biological replicates (numbered 1–3 below), that were either left untreated (left), or treated with 100 ng/mL sMPL (middle) or E6020 (right). Each treatment per replicate was assayed on an individual chip. Intensity was plotted relative to the average of the untreated controls. (B) Quadrant plot of gene induction/suppression by sMPL versus Control (x-axis) and E6020 versus Control (y-axis). Diagonal lines are spaced 1 log2-fold difference, and genes falling within the lines are equally induced by sMPL and E6020. Genes of particular interest for TLR4-induced, MyD88-dependent and TRIF-dependent activation are labeled. (C) The ratio of induction between E6020-treated cells and sMPL-treated cells was calculated for a subset of the differentially expressed genes with previously reported dependence on MyD88, TRIF, or both adapters downstream of TLR4 signaling pathways (Supplementary Table II).