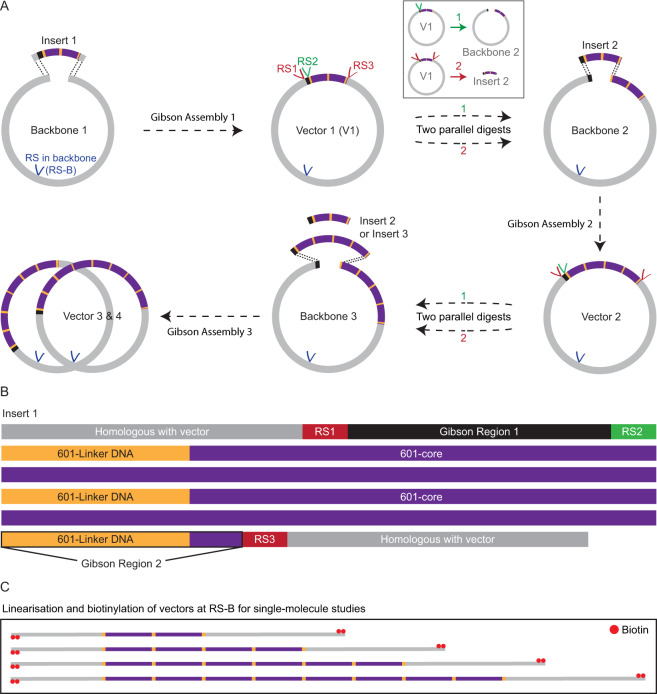
Figure 1.
Schematic representation of a Gibson Assembly based cloning strategy for obtaining arrays of 601 nucleosome positioning motifs. (A) Insertion of a segment containing n × 601 repeats into a linearised plasmid via sequential Gibson Assembly reactions. In the first Gibson Assembly reaction, a fragment containing two 601-core repeats flanked by identical linker sequences (Insert 1) is embedded in a suitable plasmid (Backbone 1). The resulting vector (Vector 1) can then be used to obtain a new insert (Insert 2) containing two 601 motifs via digestion at restriction sites RS1 and RS3 (Inset). In a parallel reaction (Inset), Vector 1 can also be digested at restriction site RS2 to yield a backbone containing two 601 repeats into which Insert 2 can be embedded via a Gibson Assembly reaction. This procedure can be repeated until the desired number of 601 motifs has been obtained. (B) Sequence composition of Insert 1. Two 601-core repeats (corresponding to the 147 base pairs of the 601-core sequence, purple) are flanked by identical linker sequences (yellow). The ends of Insert 1 (grey) are homologous with the ends of Backbone 1 to facilitate the first Gibson Assembly reaction. Additionally, Insert 1 contains two ‘Gibson regions’ (Gibson Region 1 and Gibson Region 2), as well as three restriction sites (RS1, RS2, and RS3), designed in such a way that, once Insert 1 has been incorporated into Backbone 1, further 601 motifs can be embedded via subsequent Gibson Assembly steps (as shown in panel A). (C) The library of plasmids containing n × 601 repeats prepared using the approach outlined in panel A can be used directly for single-molecule studies after linearisation and biotinylation at an appropriate restriction site (RS-B in panel A).