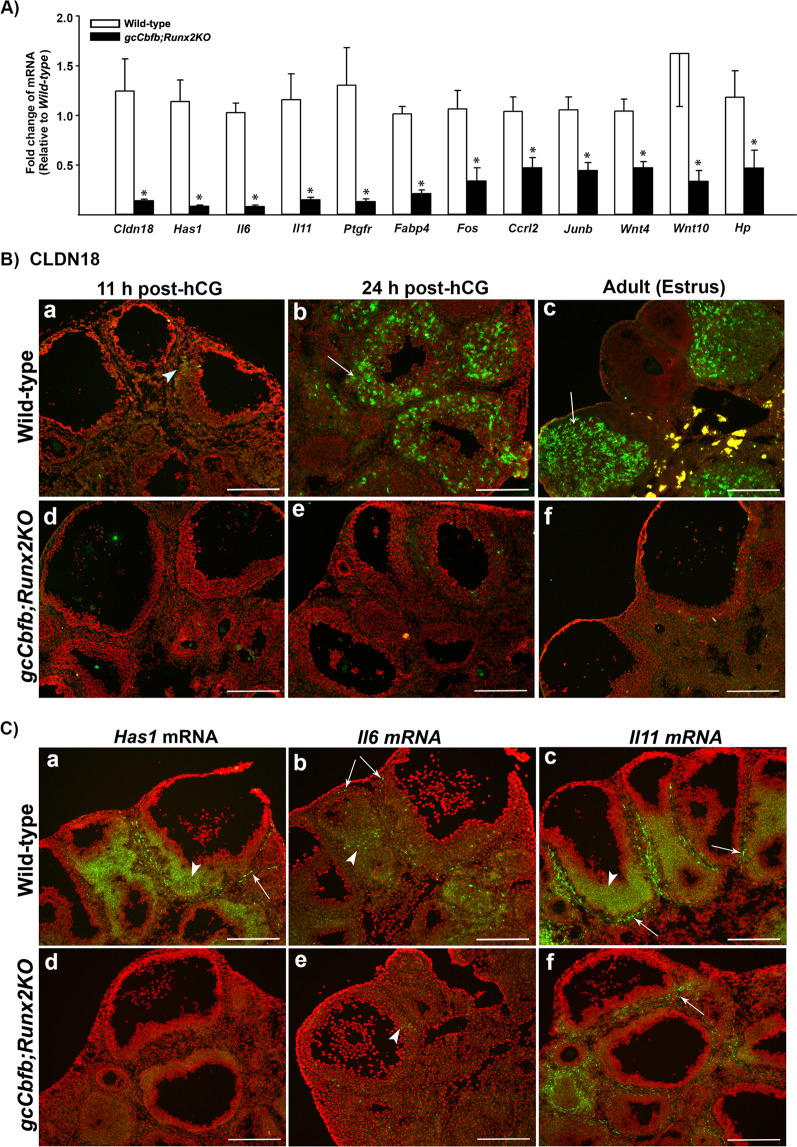
Figure 4.
Identification of down-regulated genes in ovaries of gcCbfb;Runx2KO mice (I). (A) Ovaries were collected at 11 h after hCG administration from wild-type and gcCbfb;Runx2KO mice. A list of the most highly downregulated genes was selected from RNA-seq data analyses. The levels of mRNA for genes were measured by qPCR, normalizing to the Rpl19 value in each sample (n = 8 and 7 for control and mutant mice, respectively). *p < 0.01. (B) Ovaries were collected at 11 and 24 h after hCG administration from immature animals as well as in the morning of estrus from adult mice. The localization of CLDN18 was evaluated via immunohistochemical analyses (n = 3/genotype). CLDN18 was detected as green fluorescent staining, and the tissue sections were counterstained with propidium iodide (red). Arrowheads and arrows point to a few granulosa cells and luteal cells stained positively for CLDN18, respectively. (C) in situ hybridization analyses were used to localize mRNA for Has1, IL6, and IL11 in ovaries collected at 11 h post-hCG (n = 3/genotype). Transcripts for these genes were detected as green fluorescence signals. The tissue sections were counterstained with propidium iodide (red). Arrowheads and arrows point to granulosa cells of preovulatory follicles and cells in the theca layer positive for these gene transcripts, respectively. Scale bars, 250 μm for all the images.