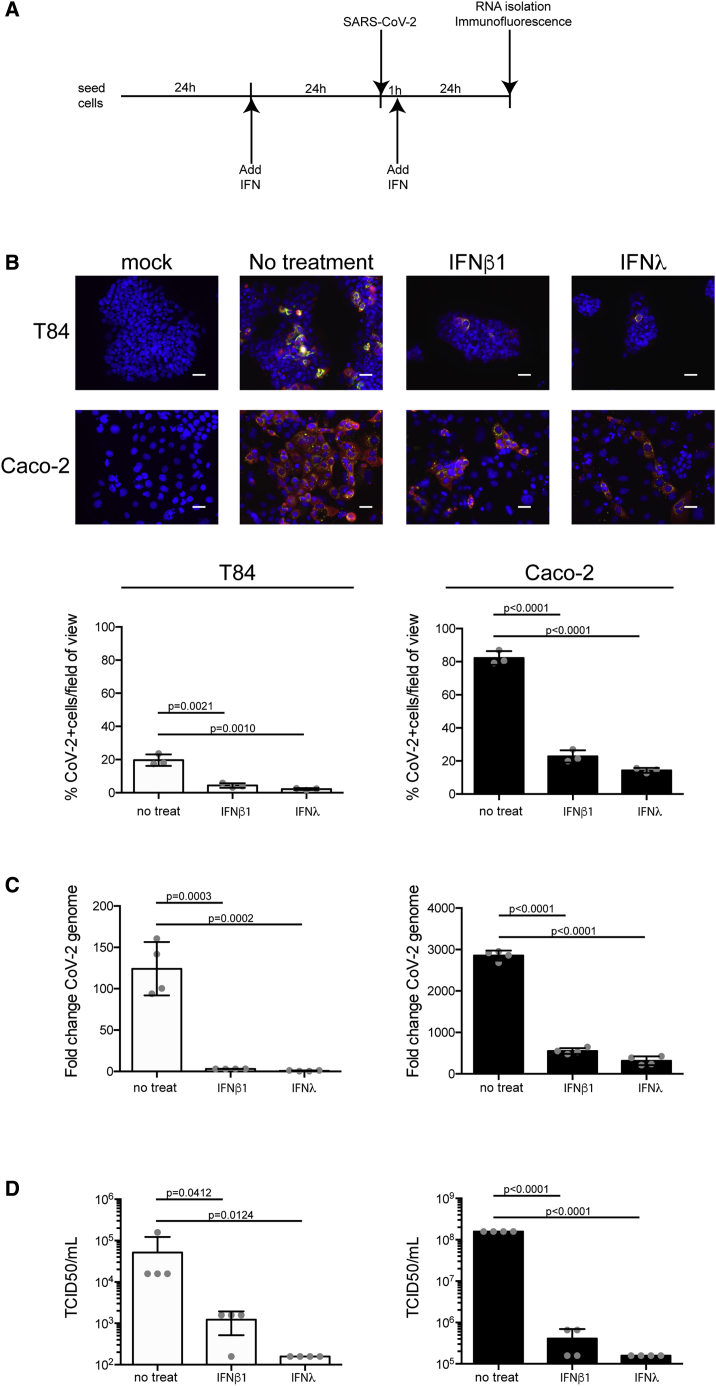
Figure 2.
IFN-β and IFN-λ Control SARS-CoV-2 Infection of hIECs
(A) Schematic describing the experimental setup.
(B) T84 and Caco-2 cells were pretreated with IFNs 24 h prior to infection. 24 hpi, virus infection was evaluated by indirect immunofluorescence for the viral N protein (red) and dsRNA (green). Nuclei were stained with DAPI (blue). Representative images are shown. The number of SARS-CoV-2-positive cells was quantified in 10 fields of view for each time point. Scale bars, 10 μm.
(C) Same as (B), except RNA was harvested and qRT-PCR was used to evaluate the copy number of SARS-CoV-2 genome.
(D) 24 hpi, supernatants were collected from infected T84 and Caco-2 cells. The amount of de novo virus present in the supernatants was determined using a TCID50 assay.
n = 3 biological replicates. Error bars indicate standard deviation.