Abstract
OBJECTIVE
Type 2 diabetes develops for many years before diagnosis. We aimed to reveal early metabolic features characterizing liability to adult disease by examining genetic liability to adult type 2 diabetes in relation to metabolomic traits across early life.
RESEARCH DESIGN AND METHODS
Up to 4,761 offspring from the Avon Longitudinal Study of Parents and Children were studied. Linear models were used to examine effects of a genetic risk score (162 variants) for adult type 2 diabetes on 229 metabolomic traits (lipoprotein subclass–specific cholesterol and triglycerides, amino acids, glycoprotein acetyls, and others) measured at age 8 years, 16 years, 18 years, and 25 years. Two-sample Mendelian randomization (MR) was also conducted using genome-wide association study data on metabolomic traits in an independent sample of 24,925 adults.
RESULTS
At age 8 years, associations were most evident for type 2 diabetes liability (per SD higher) with lower lipids in HDL subtypes (e.g., −0.03 SD [95% CI −0.06, −0.003] for total lipids in very large HDL). At 16 years, associations were stronger with preglycemic traits, including citrate and with glycoprotein acetyls (0.05 SD; 95% CI 0.01, 0.08), and at 18 years, associations were stronger with branched-chain amino acids. At 25 years, associations had strengthened with VLDL lipids and remained consistent with previously altered traits, including HDL lipids. Two-sample MR estimates among adults indicated persistent patterns of effect of disease liability.
CONCLUSIONS
Our results support perturbed HDL lipid metabolism as one of the earliest features of type 2 diabetes liability, alongside higher branched-chain amino acid and inflammatory levels. Several features are apparent in childhood as early as age 8 years, decades before the clinical onset of disease.
Introduction
Type 2 diabetes is a metabolic disease affecting >400 million people globally (1). Its incidence is driven largely by increased adiposity (2), a strong causal risk factor (3,4); yet, the difficulty of achieving and maintaining weight loss makes disease management a lifelong and expensive task (5). This is particularly problematic considering that potentially half of those living with type 2 diabetes are undiagnosed and that the future burden is expected to be greatest in lower-income countries (1). There is therefore a clear need to minimize the impact of type 2 diabetes, and this requires biological understanding of the disease at its very earliest stages.
Type 2 diabetes is typically diagnosed when blood glucose levels are ≥7 mmol/L in the fasting state or 11.1 mmol/L in the post-challenge state, or when glycated hemoglobin levels are ≥6.5% (6); yet, glucose spikes relatively late in the disease process. Repeat clinical measures from the Whitehall II cohort study suggest that insulin sensitivity starts declining a decade before glucose changes are detectable (7). Cohort studies with metabolomic measurements also observe associations of numerous subclinical traits with lower insulin sensitivity, including higher branched-chain amino acid (BCAA) concentrations, higher fatty acid and inflammatory glycoprotein concentrations, and elevated lactate and pyruvate (8–11). Relations with ketone bodies are less clear, with higher levels associated with both higher insulin sensitivity (9) and higher type 2 diabetes risk (12). Hyperglycemia also associates strongly with lipoprotein cholesterol and triglycerides (13). Whether such trait alterations reflect developmental stages of type 2 diabetes is unclear because of inherent confounding by other disease processes.
Another approach to causal inference is to examine genetic liability to type 2 diabetes in relation to metabolic traits to identify perturbations specific to the development of type 2 diabetes itself (i.e., its early metabolic features) (14). The few studies that have investigated this suggest effects of type 2 diabetes liability on cholesterol and triglycerides in HDL and VLDL particles (15), BCAAs (16,17), and ketone bodies (12). Most used a small set of genetic variants from early genome-wide association studies (GWAS), and all relied on one-off measures of metabolic traits among middle- to older-aged adults, which gives little insight about when in life metabolic alterations first occur. Examining genetic liability to type 2 diabetes in relation to repeated measures of metabolic traits starting in childhood could reveal the existence and timing of subclinical trait perturbations most central to type 2 diabetes development.
We aimed in this study to reveal early metabolic features characterizing liability to type 2 diabetes. Using birth cohort study data, we examined genetic liability to adult type 2 diabetes in relation to detailed traits from targeted metabolomics among the same individuals at four key stages of early life—childhood (age 8 years), adolescence (16 years), young adulthood (18 years), and adulthood (25 years). For replication, two-sample Mendelian randomization (MR) (18) was conducted in an independent sample of adults to confirm the persistence of any metabolic features of disease liability observed in early life.
Research Design and Methods
Study Population
Data were from offspring participants of the Avon Longitudinal Study of Parents and Children (ALSPAC), a population-based birth cohort study in which 14,541 pregnant women with an expected delivery date between 1 April 1991 and 31 December 1992 were recruited from the former Avon County of southwest England (19). Since then, 13,988 offspring alive at 1 year have been monitored repeatedly with questionnaire- and clinic-based assessments (20), with an additional 811 children enrolled over the course of the study. Offspring were considered for the current analyses if they had no older siblings in ALSPAC (202 excluded) and were of white European ethnicity (604 excluded) to reduce confounding of associations by high relatedness and ancestral population structure.
Written informed consent was provided, and ethical approval was obtained from the ALSPAC Law and Ethics Committee and the local research ethics committee. The study website contains details of all available data through a fully searchable data dictionary and variable search tool (https://www.bristol.ac.uk/alspac/researchers/our-data/).
Assessment of Genetic Liability to Adult Type 2 Diabetes
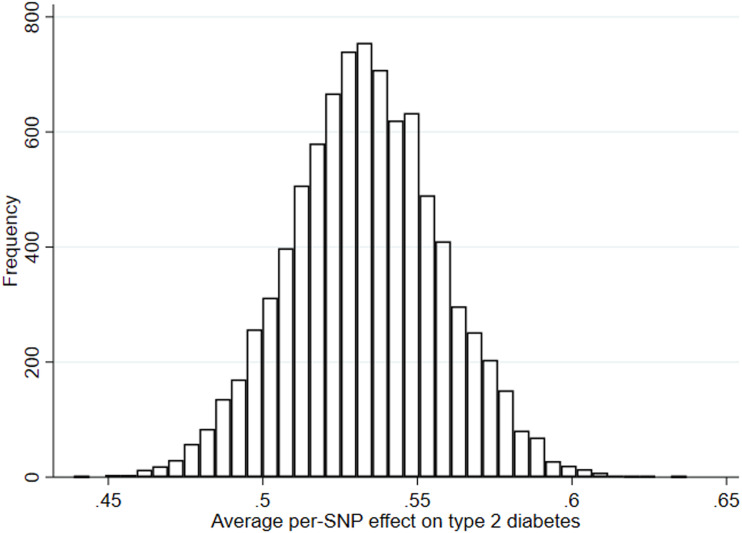
Genotype was assessed using the Illumina HumanHap550 quad chip platform. Quality control measures included exclusion of participants with sex mismatch, minimal or excessive heterozygosity, disproportionately missing data, insufficient sample replication, cryptic relatedness, and non-European ancestry. Imputation was performed using the Haplotype Reference Consortium panel. Because this study aimed to address causation, not prediction, genetic liability to type 2 diabetes was based on genetic variants associated with type 2 diabetes case status at genome-wide significance in the largest GWAS to date, which identified up to 403 independent polymorphisms among adults (74,124 case subjects and 824,006 control subjects) of white European ethnicity, explaining 17.4% of variance (21). This set of variants was refined by excluding 105 variants with P values ≥5.00 × 10−8, 12 additional variants identified only when adjusting for BMI, and variants that were in linkage disequilibrium based on R2 > 0.001 (retaining those single nucleotide polymorphisms [SNPs] with the lowest P values) using the TwoSampleMR package (22). This left 167 variants highly independently associated with adult type 2 diabetes (explaining 7.1% of variance in the UK Biobank study, using phenotyping as previously reported [21]) (Supplementary Table 1), 162 of which were available in imputed ALSPAC genotype data postquality control. This set was combined into a genetic risk score (GRS) using PLINK 1.9 (www.cog-genomics.org/plink/1.9/), specifying the effect (type 2 diabetes raising) allele and coefficient (odds ratio) from the GWAS as external weights. Scoring was done by multiplying the number of effect alleles (or probabilities of effect alleles if imputed) at each SNP (0, 1, or 2) by its weighting, summing these, and dividing by the total number of SNPs used. The score therefore reflects the average per-SNP effect on type 2 diabetes (Fig. 1).
Figure 1.
Distribution of the GRS for adult type 2 diabetes among ALSPAC offspring.
Assessment of Metabolic Traits
Participants provided nonfasting blood samples during a clinic visit while aged ∼8 years and fasting blood samples from clinic visits while aged ∼16 years, 18 years, and 25 years. Proton nuclear magnetic resonance (1H-NMR) spectroscopy from a targeted metabolomics platform (23) was performed using EDTA plasma (stored at or below −70°C preprocessing) to quantify 229 traits (149 concentrations plus 80 ratios). These included the concentration and size of lipoprotein subclass particles and their cholesterol and triglyceride content, apolipoproteins, fatty acids, preglycemic factors including lactate and glucose, amino acids, ketone bodies, and inflammatory glycoprotein acetyls.
Assessment of Adiposity and Type 2 Diabetes Status
For descriptive purposes, BMI was calculated at each time point as weight (kg) divided by height squared (m2) based on clinic measures of weight to the nearest 0.1 kg using a Tanita scale and height measured in light clothing without shoes to the nearest 0.1 cm using a Harpenden stadiometer. Type 2 diabetes/prediabetes status was not assessed at 8 years because blood glucose was not quantified based on fasting samples, and no data were collected regarding physician diagnosis. Type 2 diabetes was defined at age 16 years as fasting glucose ≥7 mmol/L and at 18 years and 25 years as fasting glucose ≥7 mmol/L or reported physician diagnosis of type 2 diabetes by that age (6). Prediabetes was defined at 16 years, 18 years, and 25 years as fasting glucose between 5.6 and 6.9 mmol/L (6).
Statistical Approach
In the first set of analyses, separate linear regression models with robust SEs were used to estimate coefficients and 95% CIs for associations of the type 2 diabetes GRS (in SD units), with each metabolic trait measured at 8 years (also in SD units) as a dependent variable adjusted for sex and age at the time of metabolic trait assessment. These were repeated for metabolic traits measured at 16 years, 18 years, and 25 years. Estimates are interpreted within a “reverse MR” framework (14), wherein the direction of causation is from disease liability to metabolic traits, and are taken to reflect “metabolic features” of type 2 diabetes liability. To compare evidence of linear change in coefficients across time points, we ran separate linear mixed models using repeated measures of each metabolic trait and examined P values from an interaction term between the type 2 diabetes GRS and the mean age at each time point (as an occasion/time variable). All models were additionally run using original (non-SD; mostly mmol/L) units to aid clinical interpretation and external comparisons.
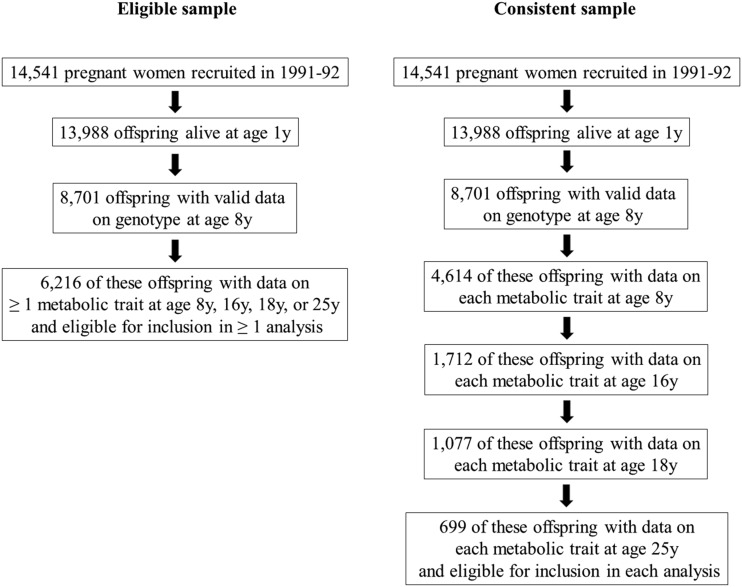
To allow full use of measured data, the aforementioned analyses were first conducted using maximum numbers of participants (N varying across ages and between traits). Participants were eligible for analyses at any age if they had data on genotype, sex, age, and at least one metabolic trait. This resulted in 6,216 eligible participants (i.e., who contributed data to any analysis), including up to 4,761, 2,928, 2,612, and 2,560 participants in models at 8 years, 16 years, 18 years, and 25 years, respectively. Analyses were repeated using a consistent (complete-case) sample of participants with data on genotype, sex, age, and all metabolic traits at each time point (Fig. 2).
Figure 2.
Selection of ALSPAC participants into analyses.
Additional Analyses
Effects of type 2 diabetes liability on measured BMI at each time point were also examined. To examine mediation of effects of type 2 diabetes liability on metabolic traits by BMI and insulin (not measured by NMR), we conducted additional analyses wherein we adjusted associations between the type 2 diabetes GRS and metabolic traits for measured BMI at the time of each trait assessment and separately for measured fasting insulin (measured using conventional clinical assays) at the time of each metabolic trait assessment (except for 8 years when insulin was not measured). We repeated these adjustments using a GRS for BMI based on 312 SNPs independently associated with adult BMI (at R2 < 0.001 and P < 5.00 × 10−8) (24) (Supplementary Table 1), 308 of which were available in the imputed ALSPAC genotype data post–quality control, and separately for a GRS for fasting insulin based on 14 SNPs independently associated with adult fasting insulin (at R2 < 0.001 and P < 5.00 × 10−8; unadjusted for BMI) (25) (Supplementary Table 1). ALSPAC analyses were conducted using Stata 15.1 software (StataCorp, College Station, TX).
We conducted two-sample MR analyses to examine metabolic features of type 2 diabetes liability using SNP outcome (metabolic trait) estimates from a GWAS of 123 traits quantified using the same NMR platform as used in ALSPAC (26) among an independent sample of 13,476–24,925 adults of European ancestry (26). Across studies included in this GWAS, mean (SD) age ranged from 23.9 (2.1) years to 61.3 (2.9) years and female sex ranged from 37 to 64%. These estimates were combined with SNP-exposure (type 2 diabetes) estimates based on the 167 SNPs for type 2 diabetes described previously (Supplementary Table 1). Three statistical methods were used to generate MR estimates using the TwoSampleMR package (22): random-effects inverse variance weighted (IVW) (22), MR-Egger, and weighted median, which make differing assumptions about directional pleiotropy (27,28). Estimates are interpreted as SD-unit differences in metabolic trait per 1-log-odds of type 2 diabetes.
This study involves describing global patterns of effect estimates; we therefore provide exact P values and focus on effect size and precision (29,30).
Data and Resource Availability
Individual-level ALSPAC data are available following an application. This process of managed access is detailed at www.bristol.ac.uk/alspac/researchers/access. Cohort details and data descriptions for ALSPAC are publicly available at the same web address. Summary-level GWAS data used in this study are publicly available without the need for application through the MR-Base platform, which is accessible at https://www.mrbase.org/.
Results
Sample Characteristics
Of the eligible participants, 49.7% were male, and BMI was higher at later ages (Table 1). Prevalence of type 2 diabetes and prediabetes was very low across time points (e.g., fewer than five cases of type 2 diabetes at 16 years, five cases (0.2%) at 18 years, and seven cases (0.4%) at 25 years). Participants who were ineligible for any analysis had slightly higher BMI than those who were eligible; type 2 diabetes prevalence and summary metabolic traits were also comparable (Supplementary Table 2).
Table 1.
Characteristics and metabolic traits at different early-life stages among 6,216 ALSPAC offspring eligible for inclusion in at least one analysis
| Childhood | Adolescence | Young adulthood | Adulthood | |
|---|---|---|---|---|
| Age (years) | 7.5 (0.3) | 15.5 (0.3) | 17.8 (0.4) | 24.5 (0.8) |
| Male, % (n) | 49.7 (3,087) | 49.7 (3,087) | 49.7 (3,087) | 49.7 (3,087) |
| BMI (kg/m2) | 16.2 (2.0) | 21.4 (3.5) | 22.7 (4.02) | 24.8 (4.9) |
| Has type 2 diabetes, % (n) | NA | NA (<5) | 0.2 (5) | 0.4 (7) |
| Has prediabetes, % (n) | NA | 0.4 (11) | 0.5 (12) | 0.4 (10) |
| Lipid traits | ||||
| Total cholesterol (mmol/L) | 3.9 (0.6) | 3.5 (0.6) | 3.5 (0.7) | 3.6 (0.8) |
| Cholesterol in VLDL (mmol/L) | 0.6 (0.2) | 0.5 (0.1) | 0.6 (0.2) | 0.4 (0.2) |
| Cholesterol in LDL (mmol/L) | 1.2 (0.3) | 1.04 (0.3) | 1.03 (0.4) | 1.2 (0.4) |
| Cholesterol in HDL (mmol/L) | 1.5 (0.2) | 1.4 (0.2) | 1.4 (0.2) | 1.4 (0.3) |
| Total triglycerides (mmol/L) | 1.1 (0.4) | 0.9 (0.3) | 0.9 (0.3) | 0.9 (0.4) |
| Triglycerides in VLDL (mmol/L) | 0.7 (0.3) | 0.6 (0.3) | 0.6 (0.3) | 0.6 (0.4) |
| Triglycerides in LDL (mmol/L) | 0.1 (0.1) | 0.1 (0.04) | 0.1 (0.1) | 0.1 (0.04) |
| Triglycerides in HDL (mmol/L) | 0.1 (0.02) | 0.1 (0.02) | 0.1 (0.02) | 0.1 (0.03) |
| Preglycemic traits | ||||
| Lactate (mmol/L) | 1.4 (0.5) | 1.3 (0.6) | 1.0 (0.5) | 0.9 (0.5) |
| Citrate (mmol/L) | 0.1 (0.03) | 0.1 (0.02) | 0.1 (0.02) | 0.2 (0.02) |
| Isoleucine (mmol/L) | 0.1 (0.02) | 0.04 (0.01) | 0.04 (0.01) | 0.1 (0.01) |
| Leucine (mmol/L) | 0.1 (0.01) | 0.1 (0.01) | 0.1 (0.01) | 0.1 (0.01) |
| Valine (mmol/L) | 0.1 (0.03) | 0.1 (0.03) | 0.1 (0.03) | 0.1 (0.03) |
| Glucose (mmol/L) | 4.2 (0.5) | 4.3 (0.3) | 4.1 (0.5) | 3.9 (0.4) |
| Inflammatory traits | ||||
| Glycoprotein acetyls (mmol/L) | 1.2 (0.1) | 1.2 (0.1) | 1.2 (0.1) | 1.2 (0.2) |
Continuous data are presented as the mean (SD) and categorical data as indicated. Type 2 diabetes is defined in adolescence as a clinic fasting glucose of ≥7 mmol/L and in young adulthood and adulthood as a clinic fasting glucose of ≥7 mmol/L or reported physician diagnosis. Prediabetes is defined as fasting glucose between 5.6 and 6.9 mmol/L. NA, not available/censored cell count.
Characteristics of the complete-case sample (N = 699) were comparable to those of the full sample (Supplementary Table 3), and differences between excluded and included participants appeared small (Supplementary Table 4).
Associations of Genetic Liability to Adult Type 2 Diabetes with Metabolic Traits at Different Early-Life Stages in ALSPAC
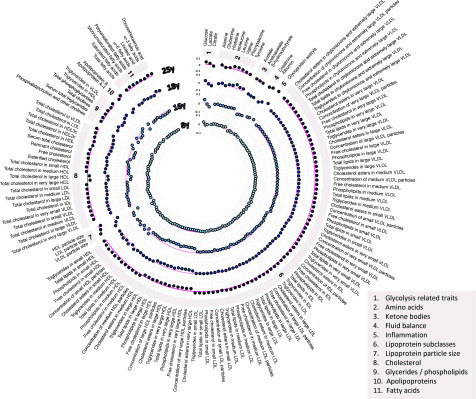
At 8 years, higher type 2 diabetes liability (per SD higher GRS) was unassociated with lipids in most lipoprotein types, including VLDL, with effects of inconsistent direction and magnitudes near zero. Associations were more consistent with lower cholesterol, triglycerides, and other lipids in very large and large HDL (e.g., −0.03 SD [95% CI −0.06, −0.003] for total lipids in very large HDL per SD higher GRS) (Fig. 3 and Supplementary Table 5).
Figure 3.
Associations of genetic liability to adult type 2 diabetes with metabolic traits at different early-life stages among ALSPAC offspring. Estimates shown are β-coefficients representing the SD difference in metabolic trait per SD higher GRS for type 2 diabetes, ordered concentrically (inner circle to outer circle) by increasing age at measurement. Six metabolic traits were not measured at the 25-years time point: diacylglycerol, ratio of diacylglycerol to triglycerides, fatty acid chain length, degree of unsaturation, conjugated linoleic acid, and ratio of conjugated linoleic acid to total fatty acids.
At 16 years, higher type 2 diabetes liability was weakly but more consistently associated with higher lipids in VLDL and lower lipids in LDL. Associations were again strongest with lower lipids in very large and large HDL (e.g., −0.08 SD [95% CI −0.11, −0.04] for total lipids in very large HDL per SD higher GRS). Preglycemic traits with the strongest evidence of association included citrate (−0.06 SD; 95% CI −0.09, −0.02) and glucose (0.05 SD; 95% CI 0.02, 0.08). Glycoprotein acetyls were associated at 0.05 SD (95% CI 0.01, 0.08).
At 18 years, higher type 2 diabetes liability remained weakly associated with higher lipids in VLDL and LDL but was more strongly associated with lower lipids in HDL, particularly very large and large HDL. Associations with BCAAs and aromatic amino acids had strengthened (e.g., valine 0.06 SD [95% CI 0.02, 0.09] and tyrosine 0.04 SD [95% CI 0.001, 0.07]). Associations remained stable with glycoprotein acetyls (0.06 SD; 95% CI 0.02, 0.10).
At 25 years, associations had strengthened between type 2 diabetes liability and lipids in VLDL subtypes such that effect sizes were comparable to those seen with lipids in HDL subtypes (e.g., 0.05 SD [95% CI 0.01, 0.09] higher total cholesterol in VLDL versus −0.06 SD [95% CI −0.09, −0.02] lower total cholesterol in very large HDL). Increasing effect size for VLDL lipids was supported by relatively low P values for trend across time points based on linear mixed models (Supplementary Table 5) (e.g., P-trend = 0.01 for total cholesterol in VLDL). These P-trend values were higher for lipids in HDL (e.g., P-trend = 0.15 for total lipids in very large HDL), indicating more stable effect sizes across time points. Associations were also more evident with several fatty acids, including a lower ratio of linoleic-to-total fatty acids (−0.07 SD; 95% CI −0.10, −0.03) and lower ratios of ω-6 to total and polyunsaturated to total fatty acids. Associations remained relatively strong with BCAAs (e.g., with leucine at 0.06 SD [95% CI 0.03, 0.10] and with glycoprotein acetyls at 0.06 SD [95% CI 0.01, 0.10]).
Association patterns were comparable when using a complete-case sample (N = 699) (Supplementary Table 6 and Supplementary Fig. 1), with estimates most consistent across time points for lipids in very large and large HDL. Results based on non-SD units for metabolic traits are in Supplementary Tables 7 and 8. Mean and SD values for metabolic traits are in Supplementary Table 9.
Associations of Genetic Liability to Adult Type 2 Diabetes with BMI at Different Early-Life Stages in ALSPAC
Type 2 diabetes liability (per SD higher GRS) was associated with higher measured BMI on each occasion. Estimates were similar to those seen for metabolic traits: 0.03 SD (95% CI 0.01, 0.05) higher BMI at 8 years, 0.05 SD (95% CI 0.02, 0.08) higher BMI at 16 years, 0.04 SD (95% CI 0.01, 0.07) higher BMI at 18 years, and 0.04 SD (95% CI 0.003, 0.07) higher BMI at 25 years. The type 2 diabetes GRS explained a low amount of variance in measured BMI (0.1% at 8 years, 0.3% at 16 years, 0.2% at 18 years, and 0.2% at 25 years).
Associations of Genetic Liability to Adult Type 2 Diabetes With Metabolic Traits at Different Early-Life Stages in ALSPAC, Adjusted for BMI and Fasting Insulin
When adjusting for measured BMI, associations between type 2 diabetes liability and metabolic traits on each occasion were largely consistent in direction and magnitude (Supplementary Table 11) (e.g., −0.05 SD [95% CI −0.08, −0.01] for lipids in very large HDL at 25 years). This was also apparent when adjusting for a GRS for BMI (Supplementary Table 12). Associations were also consistent when adjusting for measured fasting insulin on available occasions (Supplementary Table 13) (e.g., 0.06 SD [95% CI 0.03, 0.09] for leucine at 25 years) and likewise when adjusting for a GRS for fasting insulin (Supplementary Table 14) (e.g., 0.05 SD [95% CI 0.01, 0.09] for glycoprotein acetyls at 25 years).
Associations of Genetic Liability to Adult Type 2 Diabetes with Metabolic Traits in Adulthood in GWAS Summary Data
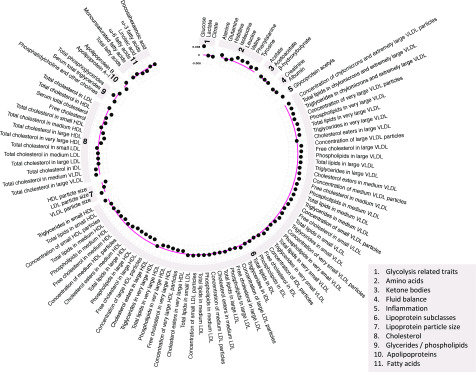
Results of two-sample MR analyses in an independent sample of adults indicated largely persistent patterns of association between genetic liability to type 2 diabetes and metabolic traits seen across early life (Fig. 4 and Supplementary Table 10). Higher genetic liability to type 2 diabetes was generally positively associated with VLDL lipid subtypes and inversely associated with HDL lipid subtypes, again for large and very large HDL specifically (e.g., −0.004 SD [95% CI −0.007, −0.002] per 1-log-odds of type 2 diabetes for total lipids in large HDL). Type 2 diabetes liability was positively associated with BCAA levels (e.g., 0.004 SD of leucine, isoleucine, and valine per 1-log-odds). There was less evidence of association between type 2 diabetes liability and glycoprotein acetyls, at 0.003 SD (95% CI 0.0001, 0.005) per 1-log-odds. The strongest association was seen for glucose, at 0.008 SD (95% CI 0.006, 0.010) per 1-log-odds. Evidence of effect heterogeneity was strong for most metabolic traits (e.g., Cochran Q P = 7.83 × 10−16 for the glucose IVW estimate). Where IVW estimates suggested evidence of effect, weighted median estimators were consistent, whereas MR-Egger estimates were imprecise, although there was little evidence to suggest that MR-Egger intercept estimates differed from zero for metabolic traits (all P > 0.003).
Figure 4.
Associations of genetic liability to adult type 2 diabetes with metabolic traits in an independent sample of adults based on two-sample MR. Estimates shown are β-coefficients representing the SD-unit difference in metabolic trait per 1-log-odds of type 2 diabetes based on the IVW method.
Conclusions
We aimed to reveal early metabolic features of type 2 diabetes liability by integrating genetic liability to adult disease with detailed metabolic traits measured across early life (from 8 to 25 years). These metabolic traits were measured long before the expected clinical onset of type 2 diabetes (31), and consequently, their perturbations are expected to reflect early signs of disease that are detectable in the circulation. Our findings suggest that one of these earliest features is lower lipid content in HDL particles, particularly in large and very large subtypes, alongside lower citrate and higher BCAA and inflammatory glycoprotein acetyl levels. Several features are apparent in childhood as early as age 8 years, several decades before the clinical onset of disease. Persistent patterns of effect were observed in an independent sample of adults using two-sample MR, supporting their continued relevance with advancing age.
Adiposity is expected to be a key driver of type 2 diabetes and its metabolic intermediates. This is supported by several MR studies suggesting strong effects of BMI on metabolic trait levels (32) and type 2 diabetes in adulthood (3,4) and by the close resemblance of the effects of adiposity to those seen presently for type 2 diabetes liability, for example, lower cholesterol in HDL, higher cholesterol in apolipoprotein-B–containing lipoproteins, higher BCAA levels, and higher glycoprotein acetyls. Presently, adult type 2 diabetes liability was found to raise BMI in childhood, with effect sizes appearing consistent at later time points and similar to those seen for metabolic traits. Adjusting metabolic effects of type 2 diabetes liability for BMI (phenotypically or genetically) produced little-to-no attenuation, however. This finding suggests that higher adiposity is one early feature of type 2 diabetes liability but that metabolic effects of liability captured by the genetic variants used here act largely independently of adiposity. This is further supported by lower variance captured in measured BMI by the type 2 diabetes GRS (0.2% at 25 years in ALSPAC) compared with the variance explained in type 2 diabetes itself (7.1% among adults in UK Biobank [21]). The type 2 diabetes GRS also explained considerably less variance in BMI than did the BMI GRS (0.2% vs. 5% at 25 years in ALSPAC, respectively).
The apparent specificity of effects of type 2 diabetes liability on lipids within large and very large HDL suggests distinct molecular functions of HDL subtypes. Medium and smaller HDLs have shown more overlapping immune-related gene expression profiles with triglyceride- and apolipoprotein-B–containing particles than have larger HDLs (33), suggesting that smaller HDLs are more functionally atherogenic, whereas larger HDLs are more functionally involved in nonatherogenic reverse cholesterol transport and more representative of conventional/nonsubtyped HDL measurements (34).
Bidirectional effects between metabolic traits and type 2 diabetes liability remain plausible. One recent MR study among adults examined effects of type 2 diabetes liability on metabolic traits derived from NMR and mass spectrometry and effects in the reverse direction for 20 traits (17). Genetically higher cholesterol in HDL (again in very large and large subtypes) was most associated with lower fasting glucose, but these effects did not extend to type 2 diabetes. Metabolic traits with the strongest evidence of effect on type 2 diabetes were phospholipids in VLDL and IDL and total triglycerides (17). Conversely, type 2 diabetes liability had the greatest effect on alanine, along with several phosphatidylcholine alkyl-acyls, supporting such perturbations as consequences rather than causes of type 2 diabetes liability.
Another MR study that instrumented BCAAs suggested that higher levels raise type 2 diabetes risk (35). Instrumenting metabolic traits is difficult, however, because their genetic architecture overlaps greatly (36,37), resulting in the use of genetic variants that are typically not specific to one metabolic trait. Using the same genetic variant for multiple traits outside of a multivariable MR framework can lead to inflated MR estimates via a “double-counting” of allele effects, as was likely the case previously (35,37) where the variant rs1440581 was used to instrument both leucine and valine. Other MR evidence suggested that higher genetic liability to insulin resistance (a type 2 diabetes precursor) raises BCAA levels (38). Strong effects of insulin resistance were also found on higher lipids in apolipoprotein-B–containing particles, on lower lipids in very large, large, and medium HDL, and on lower citrate (38)—patterns like those seen presently. Effect sizes were much larger in that previous study, likely because insulin resistance is more biologically distinct than type 2 diabetes (a heterogenous disease), with more precise metabolic effects and because of much older ages at which traits were measured. These results, together with ours, position such perturbations as consequences of type 2 diabetes development. Our results further indicate that effects of type 2 diabetes liability on metabolic traits are generally not attenuated with adjustment for fasting insulin (phenotypically or genetically), suggesting that effects of genetic liability to type 2 diabetes are potentially independent of insulin resistance. The effects of liability presently examined are based on a generalized disease phenotype, however. Genetic partitioning of type 2 diabetes into distinct subtypes may enable refined mechanistic insights.
Limitations
Limitations of this study include modest sample sizes and thus power/precision for ALSPAC estimates, particularly for complete-case analyses. Descriptive comparisons were made for key measured traits between excluded and included participants and these differences appeared small (e.g., BMI was 25.0 vs. 24.5 kg/m2 at age 25 years in the full vs. complete-case sample, respectively). Blood samples from the first occasion of metabolic trait assessment were derived while nonfasting, but trait concentrations have previously shown stability over different durations of fasting time (39). Adjustments were made for measured BMI and fasting insulin at the time of metabolic trait measurement to assess mediation via estimate attenuation. This procedure carries potential for collider bias from unmeasured confounding of the mediator-outcome association (40), but close agreement between phenotypic and genetic adjustments suggests this bias is minor.
Our analyses were restricted to white Europeans, which helps to reduce confounding by ancestral population structure but limits inference to other groups. Such inference requires more comprehensive GWAS of nonwhite European populations together with metabolomic measurements in cohort studies with higher representation of those groups. Our two-sample MR analysis confirmed the same metabolic features in an independent sample, but differences in exposure units prevent direct comparison of effect sizes. We aimed to reveal early metabolic features of type 2 diabetes, but objectives are only feasible within a framework of “liability” to type 2 diabetes because the current study population is without clinical disease. This was deliberate as part of an approach for identifying early features of disease activity in early life, with preclinical implications. Lastly, although we interpret results as reflecting metabolic effects of type 2 diabetes liability, alternative explanations, including bias or pleiotropy, remain possible (seven such scenarios are proposed [14]). Methodological flexibility together with an increasingly large scale and scope of genomic and metabolomic data should make interrogating these scenarios increasingly feasible.
Conclusion
Our results based on genetic liability to adult type 2 diabetes in relation to repeated measures of detailed metabolic traits across early life suggest that one of the earliest metabolic features of type 2 diabetes liability is lower lipid content in HDL particles—particularly in very large and large subtypes—alongside lower citrate and higher BCAA and inflammatory glycoprotein acetyl levels. Several features are apparent in childhood as early as age 8 years, several decades before the clinical onset of disease.
Article Information
Acknowledgments. The authors are grateful to the families who participated in this study, the midwives for their help in recruiting them, and the entire ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, and nurses.
Funding. The UK Medical Research Council, Wellcome (102215/2/13/2), and the University of Bristol provide core support for ALSPAC. GWAS data were generated by Sample Logistics and Genotyping Facilities at Wellcome Sanger Institute and LabCorp (Laboratory Corporation of America) using support from 23andMe. J.A.B. is supported by Cancer Research UK (C18281/A19169) and the Elizabeth Blackwell Institute for Health Research, University of Bristol, and the Wellcome Trust Institutional Strategic Support Fund (204813/Z/16/Z). C.J.B. and E.E.V. are supported by Diabetes UK (17/0005587). D.C. and G.D.S. work in a unit funded by the UK Medical Research Council (MC_UU_00011/1,6) and the University of Bristol. N.J.T. is a Wellcome Trust investigator (202802/Z/16/Z), is the principal investigator of the ALSPAC (Medical Research Council/Wellcome; 102215/2/13/2), is supported by the University of Bristol National Institute for Health Research Biomedical Research Centre (BRC-1215-20011) and the Medical Research Council Integrative Epidemiology Unit (MC_UU_12013/3), and works within the Cancer Research UK Integrative Cancer Epidemiology Programme (C18281/A19169).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Duality of Interest. As of January 2020, A.M. is an employee of Genentech and a holder of Roche stock. No other potential conflicts of interest relevant to this article were reported.
Author Contributions. J.A.B., C.J.B., M.J.G., D.C., A.M., G.D.S., N.J.T., and E.E.V. contributed to the planning of the study. J.A.B. and C.J.B. had access to data and conducted analyses. J.A.B., C.J.B., and E.E.V. wrote the first draft. All authors critically reviewed the intellectual content of manuscript drafts and approved the final version for submission. J.A.B. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Parts of this study were presented in abstract form at the 55th Annual Meeting of the European Association for the Study of Diabetes, Barcelona, Spain, 16–20 September 2019.
Footnotes
This article contains supplementary material online at https://doi.org/10.2337/figshare.12058923.
References
- 1.International Diabetes Federation IDF Diabetes Atlas, 8th ed. Brussels, Belgium, International Diabetes Federation, 2017 [Google Scholar]
- 2.Abarca-Gómez L, Abdeen ZA, Hamid ZA, et al.; NCD Risk Factor Collaboration (NCD-RisC) . Worldwide trends in body-mass index, underweight, overweight, and obesity from 1975 to 2016: a pooled analysis of 2416 population-based measurement studies in 128·9 million children, adolescents, and adults. Lancet 2017;390:2627–2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corbin LJ, Richmond RC, Wade KH, et al. . BMI as a modifiable risk factor for type 2 diabetes: refining and understanding causal estimates using Mendelian randomization. Diabetes 2016;65:3002–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dale CE, Fatemifar G, Palmer TM, et al.; UCLEB Consortium; METASTROKE Consortium . Causal associations of adiposity and body fat distribution with coronary heart disease, stroke subtypes, and type 2 diabetes mellitus: a Mendelian randomization analysis. Circulation 2017;135:2373–2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dombrowski SU, Knittle K, Avenell A, Araújo-Soares V, Sniehotta FF. Long term maintenance of weight loss with non-surgical interventions in obese adults: systematic review and meta-analyses of randomised controlled trials. BMJ 2014;348:g2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.American Diabetes Association 2. Classification and diagnosis of diabetes: Standards of Medical Care in Diabetes–2020. Diabetes Care 2020;43(Suppl. 1):S14–S31 [DOI] [PubMed] [Google Scholar]
- 7.Tabák AG, Jokela M, Akbaraly TN, Brunner EJ, Kivimäki M, Witte DR. Trajectories of glycaemia, insulin sensitivity, and insulin secretion before diagnosis of type 2 diabetes: an analysis from the Whitehall II study. Lancet 2009;373:2215–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Würtz P, Soininen P, Kangas AJ, et al. . Branched-chain and aromatic amino acids are predictors of insulin resistance in young adults. Diabetes Care 2013;36:648–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Würtz P, Mäkinen VP, Soininen P, et al. . Metabolic signatures of insulin resistance in 7,098 young adults. Diabetes 2012;61:1372–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahendran Y, Cederberg H, Vangipurapu J, et al. . Glycerol and fatty acids in serum predict the development of hyperglycemia and type 2 diabetes in Finnish men. Diabetes Care 2013;36:3732–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Würtz P, Tiainen M, Mäkinen VP, et al. . Circulating metabolite predictors of glycemia in middle-aged men and women. Diabetes Care 2012;35:1749–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahendran Y, Vangipurapu J, Cederberg H, et al. . Association of ketone body levels with hyperglycemia and type 2 diabetes in 9,398 Finnish men. Diabetes 2013;62:3618–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Stančáková A, Soininen P, et al. . Lipoprotein subclass profiles in individuals with varying degrees of glucose tolerance: a population-based study of 9399 Finnish men. J Intern Med 2012;272:562–572 [DOI] [PubMed] [Google Scholar]
- 14.Holmes MV, Davey Smith G. Can Mendelian randomization shift into reverse gear? Clin Chem 2019;65:363–366 [DOI] [PubMed] [Google Scholar]
- 15.Stančáková A, Paananen J, Soininen P, et al. . Effects of 34 risk loci for type 2 diabetes or hyperglycemia on lipoprotein subclasses and their composition in 6,580 nondiabetic Finnish men. Diabetes 2011;60:1608–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stančáková A, Civelek M, Saleem NK, et al. . Hyperglycemia and a common variant of GCKR are associated with the levels of eight amino acids in 9,369 Finnish men. Diabetes 2012;61:1895–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J, van Klinken JB, Semiz S, et al. . A Mendelian randomization study of metabolite profiles, fasting glucose, and type 2 diabetes. Diabetes 2017;66:2915–2926 [DOI] [PubMed] [Google Scholar]
- 18.Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet 2014;23:R89–R98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fraser A, Macdonald-Wallis C, Tilling K, et al. . Cohort profile: the Avon Longitudinal Study of Parents and Children: ALSPAC mothers cohort. Int J Epidemiol 2013;42:97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boyd A, Golding J, Macleod J, et al. . Cohort profile: the ‘children of the 90s’—the index offspring of the Avon Longitudinal Study of Parents and Children. Int J Epidemiol 2013;42:111–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahajan A, Taliun D, Thurner M, et al. . Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet 2018;50:1505–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hemani G, Zheng J, Elsworth B, et al. . The MR-Base platform supports systematic causal inference across the human phenome. eLife 2018;7:e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Würtz P, Kangas AJ, Soininen P, Lawlor DA, Davey Smith G, Ala-Korpela M. Quantitative serum nuclear magnetic resonance metabolomics in large-scale epidemiology: a primer on -omic technologies. Am J Epidemiol 2017;186:1084–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pulit SL, Stoneman C, Morris AP, et al.; GIANT Consortium . Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum Mol Genet 2019;28:166–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scott RA, Lagou V, Welch RP, et al.; DIAbetes Genetics Replication and Meta-analysis (DIAGRAM) Consortium . Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet 2012;44:991–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kettunen J, Demirkan A, Würtz P, et al. . Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat Commun 2016;7:11122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burgess S, Bowden J, Fall T, Ingelsson E, Thompson SG. Sensitivity analyses for robust causal inference from Mendelian randomization analyses with multiple genetic variants. Epidemiology 2017;28:30–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol 2016;40:304–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sterne JA, Davey Smith G. Sifting the evidence—what’s wrong with significance tests? BMJ 2001;322:226–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wasserstein RL, Lazar NA. The ASA’s statement on p-values: context, process, and purpose. Am Stat 2016;70:129–133 [Google Scholar]
- 31.Sharma M, Nazareth I, Petersen I. Trends in incidence, prevalence and prescribing in type 2 diabetes mellitus between 2000 and 2013 in primary care: a retrospective cohort study [published correction appears in BMJ Open 2016;6:e010210corr1]. BMJ Open 2016;6:e010210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Würtz P, Wang Q, Kangas AJ, et al. . Metabolic signatures of adiposity in young adults: Mendelian randomization analysis and effects of weight change. PLoS Med 2014;11:e1001765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inouye M, Kettunen J, Soininen P, et al. . Metabonomic, transcriptomic, and genomic variation of a population cohort. Mol Syst Biol 2010;6:441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tall AR. An overview of reverse cholesterol transport. Eur Heart J 1998;19(Suppl. A):A31–A35 [PubMed] [Google Scholar]
- 35.Lotta LA, Scott RA, Sharp SJ, et al. . Genetic predisposition to an impaired metabolism of the branched-chain amino acids and risk of type 2 diabetes: a Mendelian randomisation analysis. PLoS Med 2016;13:e1002179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shin S-Y, Fauman EB, Petersen A-K, et al.; Multiple Tissue Human Expression Resource (MuTHER) Consortium . An atlas of genetic influences on human blood metabolites. Nat Genet 2014;46:543–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holmes MV, Ala-Korpela M, Smith GD. Mendelian randomization in cardiometabolic disease: challenges in evaluating causality. Nat Rev Cardiol 2017;14:577–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Q, Holmes MV, Davey Smith G, Ala-Korpela M. Genetic support for a causal role of insulin resistance on circulating branched-chain amino acids and inflammation. Diabetes Care 2017;40:1779–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sidhu D, Naugler C. Fasting time and lipid levels in a community-based population: a cross-sectional study. Arch Intern Med 2012;172:1707–1710 [DOI] [PubMed] [Google Scholar]
- 40.Richiardi L, Bellocco R, Zugna D. Mediation analysis in epidemiology: methods, interpretation and bias. Int J Epidemiol 2013;42:1511–1519 [DOI] [PubMed] [Google Scholar]