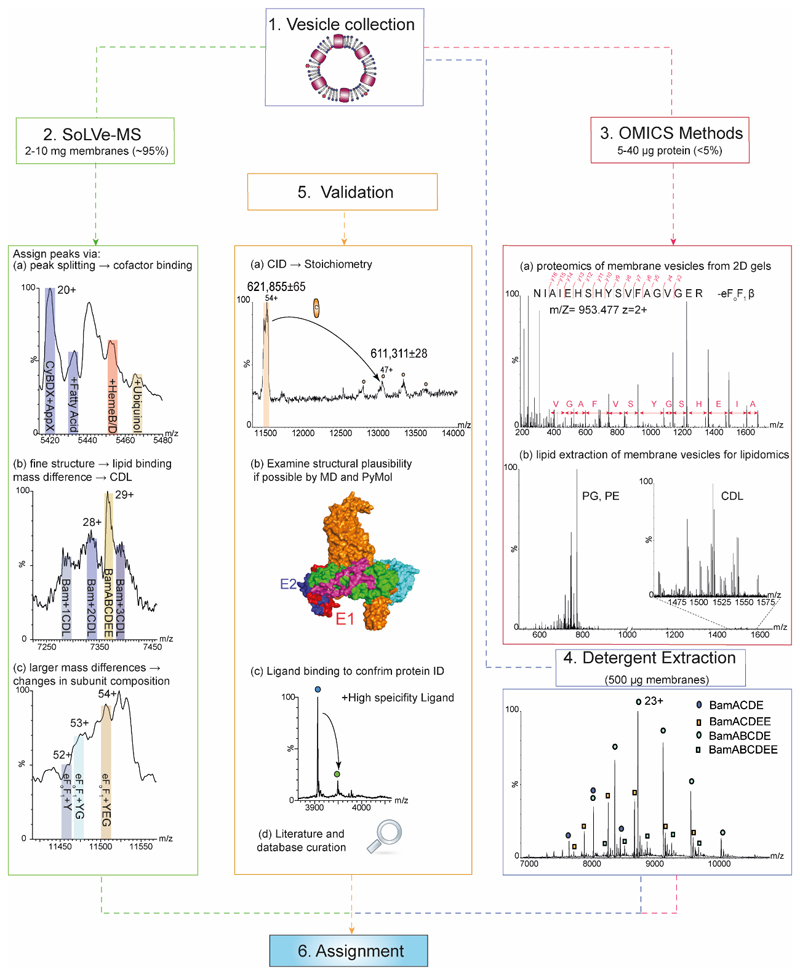
Figure 2. How to approach SoLVe-MS, data analysis and validation.
Step 1- Obtain vesicles-addressed in step 1 of Procedure section. Step 2- SoLVe-MS will require the greatest proportion of the collected vesicles (~95%). Acquire the highest resolution MS data possible and assign peaks to charge states of a protein complex. Careful examination of the spectrum is required to search for peak splitting due to cofactor binding and unique identifiers (a), or diffuse peaks indicating lipid binding and co-factors and other small molecules. (b). Unexpected large mass variation may be observed indicating differences in subunit composition (c). Step 3- Use available omics approaches. This will require μg of protein (Likely less than the remaining 5% of material). Both native and denaturing gels are needed to support interpretation. All gel bands should be excised and subjected to standard proteomics approaches leading to identification of possible candidates and their subunits. Standard proteomics shown here identifies a partial sequence of the β subunit of the eFoF1 (a). The lipid components of the preparation should be examined by lipidomics (b). Step 4- Detergent extraction of proteins directly from the same membrane as a control for stoichiometry and lipid binding, or a recombinant version of a complex, such as the Bam complex purified in C8E4 detergent can be used for comparison. Here both stoichiometries are observed (BamABCDE and a population of BamABCDEE). Step 5- When all data has been acquired, validation is undertaken. This can be done in multiple ways including literature search. A mass table derived from databases or if possible previous literature reports of measured subunit masses should be curated. Examination of structural plausibility utilizing molecular dynamics simulation or examining the structural integrity in PyMOL should be carried out. All these should eventually significantly increase the confidence in the assignment. Representative spectra are of a single repeat from 3-6 biological repeats.