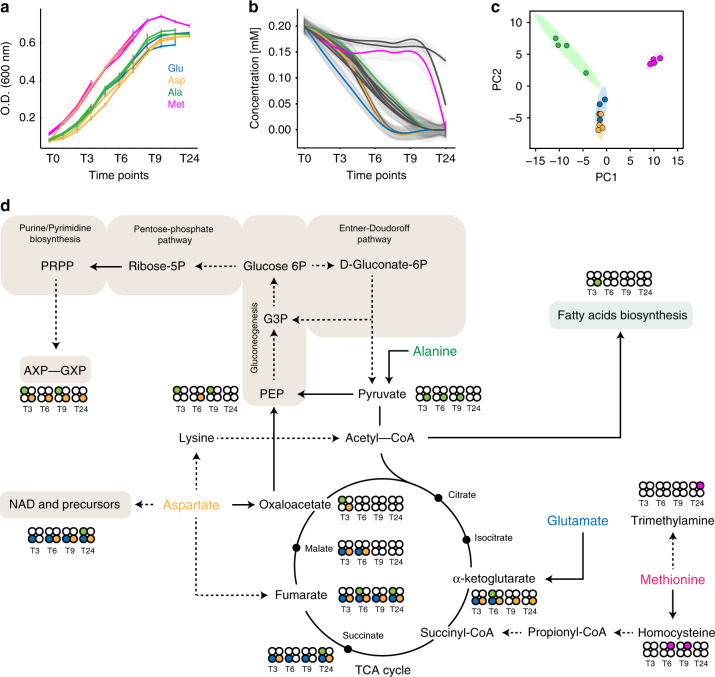
Fig. 5. The fate of catabolized amino acids.
a PhTAC125 growth curves across all replicates in the 19 AA medium with one of them being 13C labelled. Each colour indicates which was the 13C-labelled amino acids for each replicate. Error bars represent SD of four different cell cultures. b Extracellular concentration of all the amino acids in time. Colour codes as in a. c PCA on the 13C spectra. Colour codes as in a. Grey shaded area includes the 95% confidence of the linear regression (coloured) line over the concentrations of the amino acids belonging to the same group. d Central PhTAC125 metabolic network, highlighting the possible entry points of the labelled amino acids. Circles close to compounds indicate the 13C signal recovered at the different time points (T3–T24) and coming from different amino acids (colour codes as in a) corresponding. Source data are provided as a Source Data file.